Gene
KWMTBOMO16514
Pre Gene Modal
BGIBMGA009653
Annotation
PREDICTED:_transcription_factor_CP2-like_protein_1_isoform_X1_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 4.153
Sequence
CDS
ATGGAATATACGGAATATAGAGACTTTACAATGCATGACGTACGTAAAGAGGAGTACGGCTGGGAATCTACAGTTAGACTCATGGTCGGGAGGGAGGACGAATTGGGCATGTACCTGGAGGAGAGCAGGGAGGGCAGGAAGCGGAAGTGGCAGCCCGAGAAAATGAGGCGAGACGGCAGCGAAGATAAAGATCAGAAGAAGGTGCCGCCGAAGAAGATCATGGCTGCCCTAAGCGAGATAGGCGGGAGCGGCACAAGCTCCTCATCCTCGCATTTGTCTCCGGCATGGCAAGTCAACGAGCTCGATCTGGACTTACCCGTCGAACTATCCATGAACGAAGCGCTGTTGTCGCTTCCATCGCTGGCGGTGTTCAAACAGGAGGCTCCGTCTCCGACGGGCGCGGTGCTCTCGCCCCCCCGCCGCCACTGGCCCGTCCCCCGACGGAACGACGATCGGCAGATGACAAGCATGGTTGTCGACGGAGCCCGTGACGGCATGGACGAGTGCCAGCAGTCGCCTTCCCACAACGGAAACGCTATGCAGCATGGAAGTCCCGACGCCTTGCAGTGCCAACAGCAGATGCCGCCGAGCGCCGTCATGCCAATGAACGGTTACCATTCACCAAACGAACACGAGAATAAAAACGCTGGTCTGCTGATCTGCTCTCCGGTGGGCTCGCTGGAAGGCTACCTGCACTCTCCACCACACCTGCAGCAGCGTTCCGACTCCAGCTTCAAGGATGACACCAGGTTCCAGTACGTTCTGGCCGCAGCGACTTCGATAGCAACCAAACAGAACGAGGAAACCCTGACCTACCTCAACCAAGGCCAGTCCTACGAGATCAAGCTCAAGAAGCTCGGTGACTTGTCACATTACAAAGGAAAGCTGCTTAAAAGTGTCATCAAGATATGCTTCCACGAGCGCCGCTTGCAGTACATGGAAAGGGAGCAGATAGCGCAGTGGCACGCCGAGAGGCCGGGGGAGAGGATCGTAGAGGTGGACGTGCCTCTCTCGTACGGCATTATAAAAGTGGAACAATCCACAGCACTGAACGCGCTCCACGTCTACTGGGACGCTACCAAGGACGTGGGTGTTTATATAAAGGTATTCAAATTGAAAGGCGCCGATCGCAAACACAAACAGGACAGGGAGAAGGTGCTGCGCAGACCGAGGGCCGATCTGGACAAGTACCAGCCCGCCTGCGAGGCCACTGTGCTGACTACGTTGTCACATGACGCGCTGATGCCGCCGCCCTCGCTGGTCACCACCTCGCCGTCCTACTCGCCAGAGCTGTGTGTGACACCTAAATCTGCCTCGAGCCCTATCGCTACCAAGCCGATTTCTCAACCAGCCATCGTTGGTACTACTAATCAAATGAAGCCGCTGTATTGTCAAGATGCTGGTGAGATGAAGGTGAGCAGCCCAACCTGCCACTCCCCAGGAGCTATCCTGCCTGACCTACCACAGCCTGCTGAGGAACCAGACAAGAACTTTGGATTATCAAAAGATGCCACCTCCGCCGAGACTCAGGTGTGGTTGTCCCGACAACGGTTCATGCAACACGCCGCCACGTTCGCGAACTTTTCCGGGGCTGATCTGCTGAGACTATCCCGTGATGACATTATCCAGATCGTCGGACTGGCTGATGGCATACGTCTGTTCAACGCACTTCACGCCAAACGCATCTCGCCGCGCCTCACCCTGTACGTGTGCCCGCCCTCCTCCGACGTGCACTGCGCGCTCTACCTGCACGCGTGTCGCGCGCACGAGCTGCTGCACAAGCTGAACGCGCTGCAGGGAGCCGGCAGCAAGGATTGCGAAACCGTATTGGTGTCCGGTCCGAACGGCGCTCGCGTCCGCCTCACCGACGAGCTGGTGCGACATCTGCCAGACAACTCCACCTATCGCCTCAAGAGACTCGAAGACTGCAACGCTCTGCTCCTAAGCGCCACCACCGGCAACTGA
Protein
MEYTEYRDFTMHDVRKEEYGWESTVRLMVGREDELGMYLEESREGRKRKWQPEKMRRDGSEDKDQKKVPPKKIMAALSEIGGSGTSSSSSHLSPAWQVNELDLDLPVELSMNEALLSLPSLAVFKQEAPSPTGAVLSPPRRHWPVPRRNDDRQMTSMVVDGARDGMDECQQSPSHNGNAMQHGSPDALQCQQQMPPSAVMPMNGYHSPNEHENKNAGLLICSPVGSLEGYLHSPPHLQQRSDSSFKDDTRFQYVLAAATSIATKQNEETLTYLNQGQSYEIKLKKLGDLSHYKGKLLKSVIKICFHERRLQYMEREQIAQWHAERPGERIVEVDVPLSYGIIKVEQSTALNALHVYWDATKDVGVYIKVFKLKGADRKHKQDREKVLRRPRADLDKYQPACEATVLTTLSHDALMPPPSLVTTSPSYSPELCVTPKSASSPIATKPISQPAIVGTTNQMKPLYCQDAGEMKVSSPTCHSPGAILPDLPQPAEEPDKNFGLSKDATSAETQVWLSRQRFMQHAATFANFSGADLLRLSRDDIIQIVGLADGIRLFNALHAKRISPRLTLYVCPPSSDVHCALYLHACRAHELLHKLNALQGAGSKDCETVLVSGPNGARVRLTDELVRHLPDNSTYRLKRLEDCNALLLSATTGN
Summary
Uniprot
A0A139WF83
A0A1Y1LDP9
A0A2J7Q2S2
A0A1Y1LA11
A0A2J7Q2R8
A0A2J7Q2S9
+ More
V5G4Q8 A0A1L8DIC0 A0A1L8DI50 A0A1L8DIH0 A0A1L8DI60 V9I9A9 A0A1B6DG69 A0A1B6GEK0 V9I758 V9I8E3 A0A0J7L5F1 A0A232FAA7 A0A1B0CIF9 A0A158N974 A0A0P5A5H3 A0A0P5UBK3 A0A0P5TE66 A0A0P5U7Q5 A0A0P5A7Y5 A0A0P4YWA4 A0A0P5Q9F6 A0A0P4Y750 A0A0P4YGR5 A0A0P5CPX5 A0A0P4YDM2 A0A0P4Z5N9 A0A0P5UUP0 A0A0P5QLS9 A0A0P5QBR1 A0A0N8C9T4 A0A0P6ESM4 A0A0P5YUI8 A0A0P5L2F6 A0A0N8C2T6 A0A0P4YVJ3 A0A0N7ZV02 A0A0P5Y071 A0A0P6FZ32 A0A0P5SPN8 A0A0P5C4Y4 A0A0N8A0X1 A0A0P4ZLR3 A0A0P5VMZ2 A0A0P6GJK9 A0A0P5UBK1 A0A0P6JK82 A0A0P4ZDR9 A0A0N8DKE3 A0A0P5XWL5 A0A0P5BDU5 A0A0P5RZF1 A0A0V0G599 A0A0P5B1J2 A0A224X9T8 A0A1J1IB54 A0A0A9Z1B5 A0A0A9Z2U4 A0A2R5LGD9
V5G4Q8 A0A1L8DIC0 A0A1L8DI50 A0A1L8DIH0 A0A1L8DI60 V9I9A9 A0A1B6DG69 A0A1B6GEK0 V9I758 V9I8E3 A0A0J7L5F1 A0A232FAA7 A0A1B0CIF9 A0A158N974 A0A0P5A5H3 A0A0P5UBK3 A0A0P5TE66 A0A0P5U7Q5 A0A0P5A7Y5 A0A0P4YWA4 A0A0P5Q9F6 A0A0P4Y750 A0A0P4YGR5 A0A0P5CPX5 A0A0P4YDM2 A0A0P4Z5N9 A0A0P5UUP0 A0A0P5QLS9 A0A0P5QBR1 A0A0N8C9T4 A0A0P6ESM4 A0A0P5YUI8 A0A0P5L2F6 A0A0N8C2T6 A0A0P4YVJ3 A0A0N7ZV02 A0A0P5Y071 A0A0P6FZ32 A0A0P5SPN8 A0A0P5C4Y4 A0A0N8A0X1 A0A0P4ZLR3 A0A0P5VMZ2 A0A0P6GJK9 A0A0P5UBK1 A0A0P6JK82 A0A0P4ZDR9 A0A0N8DKE3 A0A0P5XWL5 A0A0P5BDU5 A0A0P5RZF1 A0A0V0G599 A0A0P5B1J2 A0A224X9T8 A0A1J1IB54 A0A0A9Z1B5 A0A0A9Z2U4 A0A2R5LGD9
EMBL
KQ971354
KYB26457.1
GEZM01062471
GEZM01062470
JAV69706.1
NEVH01019077
+ More
PNF22878.1 GEZM01062469 JAV69708.1 PNF22880.1 PNF22879.1 GALX01003456 JAB65010.1 GFDF01007959 JAV06125.1 GFDF01007958 JAV06126.1 GFDF01007960 JAV06124.1 GFDF01007961 JAV06123.1 JR036111 AEY56924.1 GEDC01014135 GEDC01012621 GEDC01010142 JAS23163.1 JAS24677.1 JAS27156.1 GECZ01013467 GECZ01008897 GECZ01006525 GECZ01005691 JAS56302.1 JAS60872.1 JAS63244.1 JAS64078.1 JR036104 AEY56920.1 JR036108 AEY56922.1 LBMM01000662 KMQ97851.1 NNAY01000542 OXU27781.1 AJWK01013295 AJWK01013296 AJWK01013297 AJWK01013298 AJWK01013299 AJWK01013300 AJWK01013301 AJWK01013302 AJWK01013303 AJWK01013304 ADTU01008785 ADTU01008786 ADTU01008787 ADTU01008788 ADTU01008789 GDIP01203775 JAJ19627.1 GDIP01115378 JAL88336.1 GDIP01128721 JAL74993.1 GDIP01116868 JAL86846.1 GDIP01215164 JAJ08238.1 GDIP01222714 JAJ00688.1 GDIQ01120372 JAL31354.1 GDIP01231716 JAI91685.1 GDIP01230410 JAI92991.1 GDIP01183118 JAJ40284.1 GDIP01231717 JAI91684.1 GDIP01217563 JAJ05839.1 GDIP01111333 GDIP01052153 LRGB01000918 JAL92381.1 KZS15144.1 GDIQ01118567 JAL33159.1 GDIQ01122974 JAL28752.1 GDIQ01100787 JAL50939.1 GDIQ01058494 JAN36243.1 GDIP01053298 JAM50417.1 GDIQ01177245 GDIQ01055845 GDIQ01055844 JAK74480.1 GDIQ01120371 JAL31355.1 GDIP01221630 JAJ01772.1 GDIP01209921 JAJ13481.1 GDIP01064744 JAM38971.1 GDIQ01051999 JAN42738.1 GDIP01137132 JAL66582.1 GDIP01191219 JAJ32183.1 GDIP01193369 JAJ30033.1 GDIP01215163 JAJ08239.1 GDIP01097842 JAM05873.1 GDIQ01051998 JAN42739.1 GDIP01115379 JAL88335.1 GDIQ01016597 JAN78140.1 GDIP01215162 JAJ08240.1 GDIP01025103 JAM78612.1 GDIP01078930 JAM24785.1 GDIP01191218 JAJ32184.1 GDIQ01112016 JAL39710.1 GECL01002910 JAP03214.1 GDIP01191220 JAJ32182.1 GFTR01007341 JAW09085.1 CVRI01000042 CRK95673.1 GBHO01035091 GBHO01005953 GBHO01005947 GBRD01007938 GBRD01007937 JAG08513.1 JAG37651.1 JAG37657.1 JAG57883.1 GBHO01005951 GBHO01005949 GBHO01005948 GBRD01007941 GBRD01007940 JAG37653.1 JAG37655.1 JAG37656.1 JAG57880.1 GGLE01004261 MBY08387.1
PNF22878.1 GEZM01062469 JAV69708.1 PNF22880.1 PNF22879.1 GALX01003456 JAB65010.1 GFDF01007959 JAV06125.1 GFDF01007958 JAV06126.1 GFDF01007960 JAV06124.1 GFDF01007961 JAV06123.1 JR036111 AEY56924.1 GEDC01014135 GEDC01012621 GEDC01010142 JAS23163.1 JAS24677.1 JAS27156.1 GECZ01013467 GECZ01008897 GECZ01006525 GECZ01005691 JAS56302.1 JAS60872.1 JAS63244.1 JAS64078.1 JR036104 AEY56920.1 JR036108 AEY56922.1 LBMM01000662 KMQ97851.1 NNAY01000542 OXU27781.1 AJWK01013295 AJWK01013296 AJWK01013297 AJWK01013298 AJWK01013299 AJWK01013300 AJWK01013301 AJWK01013302 AJWK01013303 AJWK01013304 ADTU01008785 ADTU01008786 ADTU01008787 ADTU01008788 ADTU01008789 GDIP01203775 JAJ19627.1 GDIP01115378 JAL88336.1 GDIP01128721 JAL74993.1 GDIP01116868 JAL86846.1 GDIP01215164 JAJ08238.1 GDIP01222714 JAJ00688.1 GDIQ01120372 JAL31354.1 GDIP01231716 JAI91685.1 GDIP01230410 JAI92991.1 GDIP01183118 JAJ40284.1 GDIP01231717 JAI91684.1 GDIP01217563 JAJ05839.1 GDIP01111333 GDIP01052153 LRGB01000918 JAL92381.1 KZS15144.1 GDIQ01118567 JAL33159.1 GDIQ01122974 JAL28752.1 GDIQ01100787 JAL50939.1 GDIQ01058494 JAN36243.1 GDIP01053298 JAM50417.1 GDIQ01177245 GDIQ01055845 GDIQ01055844 JAK74480.1 GDIQ01120371 JAL31355.1 GDIP01221630 JAJ01772.1 GDIP01209921 JAJ13481.1 GDIP01064744 JAM38971.1 GDIQ01051999 JAN42738.1 GDIP01137132 JAL66582.1 GDIP01191219 JAJ32183.1 GDIP01193369 JAJ30033.1 GDIP01215163 JAJ08239.1 GDIP01097842 JAM05873.1 GDIQ01051998 JAN42739.1 GDIP01115379 JAL88335.1 GDIQ01016597 JAN78140.1 GDIP01215162 JAJ08240.1 GDIP01025103 JAM78612.1 GDIP01078930 JAM24785.1 GDIP01191218 JAJ32184.1 GDIQ01112016 JAL39710.1 GECL01002910 JAP03214.1 GDIP01191220 JAJ32182.1 GFTR01007341 JAW09085.1 CVRI01000042 CRK95673.1 GBHO01035091 GBHO01005953 GBHO01005947 GBRD01007938 GBRD01007937 JAG08513.1 JAG37651.1 JAG37657.1 JAG57883.1 GBHO01005951 GBHO01005949 GBHO01005948 GBRD01007941 GBRD01007940 JAG37653.1 JAG37655.1 JAG37656.1 JAG57880.1 GGLE01004261 MBY08387.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
A0A139WF83
A0A1Y1LDP9
A0A2J7Q2S2
A0A1Y1LA11
A0A2J7Q2R8
A0A2J7Q2S9
+ More
V5G4Q8 A0A1L8DIC0 A0A1L8DI50 A0A1L8DIH0 A0A1L8DI60 V9I9A9 A0A1B6DG69 A0A1B6GEK0 V9I758 V9I8E3 A0A0J7L5F1 A0A232FAA7 A0A1B0CIF9 A0A158N974 A0A0P5A5H3 A0A0P5UBK3 A0A0P5TE66 A0A0P5U7Q5 A0A0P5A7Y5 A0A0P4YWA4 A0A0P5Q9F6 A0A0P4Y750 A0A0P4YGR5 A0A0P5CPX5 A0A0P4YDM2 A0A0P4Z5N9 A0A0P5UUP0 A0A0P5QLS9 A0A0P5QBR1 A0A0N8C9T4 A0A0P6ESM4 A0A0P5YUI8 A0A0P5L2F6 A0A0N8C2T6 A0A0P4YVJ3 A0A0N7ZV02 A0A0P5Y071 A0A0P6FZ32 A0A0P5SPN8 A0A0P5C4Y4 A0A0N8A0X1 A0A0P4ZLR3 A0A0P5VMZ2 A0A0P6GJK9 A0A0P5UBK1 A0A0P6JK82 A0A0P4ZDR9 A0A0N8DKE3 A0A0P5XWL5 A0A0P5BDU5 A0A0P5RZF1 A0A0V0G599 A0A0P5B1J2 A0A224X9T8 A0A1J1IB54 A0A0A9Z1B5 A0A0A9Z2U4 A0A2R5LGD9
V5G4Q8 A0A1L8DIC0 A0A1L8DI50 A0A1L8DIH0 A0A1L8DI60 V9I9A9 A0A1B6DG69 A0A1B6GEK0 V9I758 V9I8E3 A0A0J7L5F1 A0A232FAA7 A0A1B0CIF9 A0A158N974 A0A0P5A5H3 A0A0P5UBK3 A0A0P5TE66 A0A0P5U7Q5 A0A0P5A7Y5 A0A0P4YWA4 A0A0P5Q9F6 A0A0P4Y750 A0A0P4YGR5 A0A0P5CPX5 A0A0P4YDM2 A0A0P4Z5N9 A0A0P5UUP0 A0A0P5QLS9 A0A0P5QBR1 A0A0N8C9T4 A0A0P6ESM4 A0A0P5YUI8 A0A0P5L2F6 A0A0N8C2T6 A0A0P4YVJ3 A0A0N7ZV02 A0A0P5Y071 A0A0P6FZ32 A0A0P5SPN8 A0A0P5C4Y4 A0A0N8A0X1 A0A0P4ZLR3 A0A0P5VMZ2 A0A0P6GJK9 A0A0P5UBK1 A0A0P6JK82 A0A0P4ZDR9 A0A0N8DKE3 A0A0P5XWL5 A0A0P5BDU5 A0A0P5RZF1 A0A0V0G599 A0A0P5B1J2 A0A224X9T8 A0A1J1IB54 A0A0A9Z1B5 A0A0A9Z2U4 A0A2R5LGD9
PDB
5MR7
E-value=3.92738e-09,
Score=149
Ontologies
PANTHER
Topology
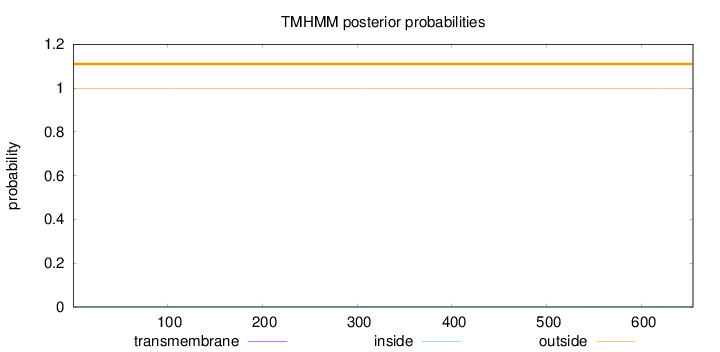
Length:
654
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00471
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00086
outside
1 - 654
Population Genetic Test Statistics
Pi
2.531975
Theta
21.371832
Tajima's D
-1.752018
CLR
0.001902
CSRT
0.0313484325783711
Interpretation
Uncertain
