Gene
KWMTBOMO16496
Annotation
S08404_hypothetical_protein_1_-_silkworm_transposon_mag
Location in the cell
Nuclear Reliability : 2.696
Sequence
CDS
ATGGGTGAGGAAGCGTACGATTTATTATCGACATTAGCCAGCCCTTCGAAGCCGTCACAATTAACATATGCTAATGCTGTAGCGATGTTGGCAGCTCACCTGCAACCGAAGCCCTCAATCTTGGCGGAAAGATACAAGTTTCGACAACGTCGGCAGCTGAATGAGTCAATAGCTGACTACCTCACAGAATTAAAAAAAATATCAAAAAATTGTGAATTCGGCTCGTCGCTGGATGAAAATCTGCGTGATCAAATGGTTTGTGGATTAAAGAGCGAAATCATACGGCAAAGGTTATTTGCCGAGGAGAAATTAGAATATAAGCGTGCAGTTACGTTAGCTTTGTCATTAGAAGCTGCGGAGAGGGATGCGATTGCAGTGGAACGTACGCCAATTGAGGAGGTTAACAAAATTAACTTCAATGAATGTTCAAGATGTGGAGACAGGAGACACCAAGCAAAGGATTGCATATACAAAGACTACGTGTGCAGTTCATGTCATGAAACCGGCCATCTACGAAGAATGTGCCCCAAAAACGGGCTTAAGAACCAGGCGGAGGCAGCCGGGGGTTCAGCACGCACCGGGCGCCGGGGAACCGGGGTGCAGGCGGCAACAGGCGATCGTGGCGCGGGCAGCGCGGTGCGGGACGAGGAGGCCGAGAGCGGGCTCCTTTGCACTTATTATACGATCAAAACAACGATGACACATACAACAACTTCAATAATGAACAGGACGGGTGTGGAGAAGAAAATGAGCCTATGTACCAAATGA
Protein
MGEEAYDLLSTLASPSKPSQLTYANAVAMLAAHLQPKPSILAERYKFRQRRQLNESIADYLTELKKISKNCEFGSSLDENLRDQMVCGLKSEIIRQRLFAEEKLEYKRAVTLALSLEAAERDAIAVERTPIEEVNKINFNECSRCGDRRHQAKDCIYKDYVCSSCHETGHLRRMCPKNGLKNQAEAAGGSARTGRRGTGVQAATGDRGAGSAVRDEEAESGLLCTYYTIKTTMTHTTTSIMNRTGVEKKMSLCTK
Summary
Similarity
Belongs to the complex I LYR family.
Uniprot
A0A3S2PB62
A0A0L7L8L8
A0A2B4RTJ4
A0A3B3SNE8
A0A2B4SPW6
A0A2S2PAH0
+ More
A0A3P8NG55 A0A1Y1LPV1 A0A1A8C840 A0A3S3NQ29 A0A1B6CPB9 A0A1B6DD07 K7EWQ6 A0A1Y1MRA2 A0A1E1XFV8 A0A3B3T0U0 A0A3Q1EQC9 A0A1A8FCH6 A0A3P8NEQ7 A0A2B4SG71 A0A2B4RPN6 A0A147BCB7 A0A2A4K2Y4 L7MMK9 A0A131XQT5 A0A1B6CK52 A0A1B6E6W0 A0A0S7IPT0 A0A1A8V6A3 W4LLX0 A0A1B6MJ81 A0A3P8VLQ9 A0A0L7L4A5 A0A1B6MUT2 A0A0L7KTF6 X1XU28 A0A3B3H2N7 A0A3P8VSE7 A0A2B4SFB0 L7MC08 A0A3B3H285 A0A2B4RXD0 J9LCT9 A0A2H1V4Y6 A0A3P8Q506 A0A085N6E6 A0A2S2PU23 A0A1B6L0V9 A0A1B6KDA3 A0A2G8KZ95 A0A0L7K286 A0A293MR82 A0A1Z5KXP3 A0A182X639 A0A085LNM9 A0A2B4T0D2 A0A2I4C6V0 A0A3N0YT70 A0A2R5LIR9 A0A0L7K3M9 A0A3B4VPK4 A0A0L7L085 A0A1B8XTH5 A0A3R7GU59 A0A0L7LC80 A0A3B3XP10 A0A1Y1M058 A0A3S0ZV35 A0A1W7R676 A0A1W7R6B8 A0A3B3Q7L2 A2TGR4 A0A085MRV2 W4LLJ5 A0A131YR50 A0A2B4R781 A0A085N4P6
A0A3P8NG55 A0A1Y1LPV1 A0A1A8C840 A0A3S3NQ29 A0A1B6CPB9 A0A1B6DD07 K7EWQ6 A0A1Y1MRA2 A0A1E1XFV8 A0A3B3T0U0 A0A3Q1EQC9 A0A1A8FCH6 A0A3P8NEQ7 A0A2B4SG71 A0A2B4RPN6 A0A147BCB7 A0A2A4K2Y4 L7MMK9 A0A131XQT5 A0A1B6CK52 A0A1B6E6W0 A0A0S7IPT0 A0A1A8V6A3 W4LLX0 A0A1B6MJ81 A0A3P8VLQ9 A0A0L7L4A5 A0A1B6MUT2 A0A0L7KTF6 X1XU28 A0A3B3H2N7 A0A3P8VSE7 A0A2B4SFB0 L7MC08 A0A3B3H285 A0A2B4RXD0 J9LCT9 A0A2H1V4Y6 A0A3P8Q506 A0A085N6E6 A0A2S2PU23 A0A1B6L0V9 A0A1B6KDA3 A0A2G8KZ95 A0A0L7K286 A0A293MR82 A0A1Z5KXP3 A0A182X639 A0A085LNM9 A0A2B4T0D2 A0A2I4C6V0 A0A3N0YT70 A0A2R5LIR9 A0A0L7K3M9 A0A3B4VPK4 A0A0L7L085 A0A1B8XTH5 A0A3R7GU59 A0A0L7LC80 A0A3B3XP10 A0A1Y1M058 A0A3S0ZV35 A0A1W7R676 A0A1W7R6B8 A0A3B3Q7L2 A2TGR4 A0A085MRV2 W4LLJ5 A0A131YR50 A0A2B4R781 A0A085N4P6
Pubmed
EMBL
RSAL01000130
RVE46453.1
JTDY01002326
KOB71659.1
LSMT01000298
PFX20931.1
+ More
LSMT01000050 PFX30415.1 GGMR01013781 MBY26400.1 GEZM01051399 JAV74971.1 HADZ01011823 HAEA01016148 SBP75764.1 NCKU01014527 RWR99571.1 GEDC01021971 JAS15327.1 GEDC01013717 JAS23581.1 AGCU01195578 GEZM01030100 JAV85807.1 GFAC01001043 JAT98145.1 HAEB01010230 SBQ56757.1 LSMT01000091 PFX28043.1 LSMT01000354 PFX19571.1 GEGO01007419 JAR87985.1 NWSH01000218 PCG78288.1 GACK01000385 JAA64649.1 GEFM01006397 JAP69399.1 GEDC01023506 GEDC01023205 JAS13792.1 JAS14093.1 GEDC01007675 GEDC01003661 JAS29623.1 JAS33637.1 GBYX01426815 JAO54450.1 HAEJ01014645 SBS55102.1 AZHX01001899 ETW98899.1 GEBQ01003960 JAT36017.1 JTDY01003118 KOB70119.1 GEBQ01006635 GEBQ01000281 JAT33342.1 JAT39696.1 JTDY01006045 KOB66341.1 ABLF02019752 LSMT01000102 PFX27500.1 GACK01003652 JAA61382.1 LSMT01000299 PFX20902.1 ABLF02015736 ABLF02039528 ABLF02042142 ODYU01000701 SOQ35910.1 KL367545 KFD65042.1 GGMR01020219 MBY32838.1 GEBQ01022644 GEBQ01012433 JAT17333.1 JAT27544.1 GEBQ01030602 JAT09375.1 MRZV01000292 PIK53317.1 JTDY01015222 KOB51965.1 GFWV01017841 MAA42569.1 GFJQ02007084 JAV99885.1 KL363363 KFD46575.1 LSMT01000001 PFX35006.1 RJVU01027767 ROL48908.1 GGLE01005288 MBY09414.1 JTDY01012655 KOB52243.1 JTDY01004038 KOB68679.1 KV463122 OCA13964.1 NIRI01002335 RJW62347.1 JTDY01001749 KOB72999.1 GEZM01043158 JAV79264.1 RQTK01000238 RUS83602.1 GEHC01000976 JAV46669.1 GEHC01000975 JAV46670.1 EF199621 ABM90392.1 KL367711 KFD59948.1 AZHX01001887 ETW98968.1 GEDV01006828 JAP81729.1 LSMT01001234 PFX12679.1 KL367556 KFD64442.1
LSMT01000050 PFX30415.1 GGMR01013781 MBY26400.1 GEZM01051399 JAV74971.1 HADZ01011823 HAEA01016148 SBP75764.1 NCKU01014527 RWR99571.1 GEDC01021971 JAS15327.1 GEDC01013717 JAS23581.1 AGCU01195578 GEZM01030100 JAV85807.1 GFAC01001043 JAT98145.1 HAEB01010230 SBQ56757.1 LSMT01000091 PFX28043.1 LSMT01000354 PFX19571.1 GEGO01007419 JAR87985.1 NWSH01000218 PCG78288.1 GACK01000385 JAA64649.1 GEFM01006397 JAP69399.1 GEDC01023506 GEDC01023205 JAS13792.1 JAS14093.1 GEDC01007675 GEDC01003661 JAS29623.1 JAS33637.1 GBYX01426815 JAO54450.1 HAEJ01014645 SBS55102.1 AZHX01001899 ETW98899.1 GEBQ01003960 JAT36017.1 JTDY01003118 KOB70119.1 GEBQ01006635 GEBQ01000281 JAT33342.1 JAT39696.1 JTDY01006045 KOB66341.1 ABLF02019752 LSMT01000102 PFX27500.1 GACK01003652 JAA61382.1 LSMT01000299 PFX20902.1 ABLF02015736 ABLF02039528 ABLF02042142 ODYU01000701 SOQ35910.1 KL367545 KFD65042.1 GGMR01020219 MBY32838.1 GEBQ01022644 GEBQ01012433 JAT17333.1 JAT27544.1 GEBQ01030602 JAT09375.1 MRZV01000292 PIK53317.1 JTDY01015222 KOB51965.1 GFWV01017841 MAA42569.1 GFJQ02007084 JAV99885.1 KL363363 KFD46575.1 LSMT01000001 PFX35006.1 RJVU01027767 ROL48908.1 GGLE01005288 MBY09414.1 JTDY01012655 KOB52243.1 JTDY01004038 KOB68679.1 KV463122 OCA13964.1 NIRI01002335 RJW62347.1 JTDY01001749 KOB72999.1 GEZM01043158 JAV79264.1 RQTK01000238 RUS83602.1 GEHC01000976 JAV46669.1 GEHC01000975 JAV46670.1 EF199621 ABM90392.1 KL367711 KFD59948.1 AZHX01001887 ETW98968.1 GEDV01006828 JAP81729.1 LSMT01001234 PFX12679.1 KL367556 KFD64442.1
Proteomes
PRIDE
Pfam
Interpro
IPR001584
Integrase_cat-core
+ More
IPR041577 RT_RNaseH_2
IPR012337 RNaseH-like_sf
IPR021109 Peptidase_aspartic_dom_sf
IPR001878 Znf_CCHC
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR000477 RT_dom
IPR036875 Znf_CCHC_sf
IPR008011 Complex1_LYR
IPR005162 Retrotrans_gag_dom
IPR001995 Peptidase_A2_cat
IPR041373 RT_RNaseH
IPR002110 Ankyrin_rpt
IPR013520 Exonuclease_RNaseT/DNA_pol3
IPR020683 Ankyrin_rpt-contain_dom
IPR036770 Ankyrin_rpt-contain_sf
IPR001969 Aspartic_peptidase_AS
IPR005312 DUF1759
IPR018061 Retropepsins
IPR034128 K02A2.6-like
IPR041577 RT_RNaseH_2
IPR012337 RNaseH-like_sf
IPR021109 Peptidase_aspartic_dom_sf
IPR001878 Znf_CCHC
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR000477 RT_dom
IPR036875 Znf_CCHC_sf
IPR008011 Complex1_LYR
IPR005162 Retrotrans_gag_dom
IPR001995 Peptidase_A2_cat
IPR041373 RT_RNaseH
IPR002110 Ankyrin_rpt
IPR013520 Exonuclease_RNaseT/DNA_pol3
IPR020683 Ankyrin_rpt-contain_dom
IPR036770 Ankyrin_rpt-contain_sf
IPR001969 Aspartic_peptidase_AS
IPR005312 DUF1759
IPR018061 Retropepsins
IPR034128 K02A2.6-like
Gene 3D
ProteinModelPortal
A0A3S2PB62
A0A0L7L8L8
A0A2B4RTJ4
A0A3B3SNE8
A0A2B4SPW6
A0A2S2PAH0
+ More
A0A3P8NG55 A0A1Y1LPV1 A0A1A8C840 A0A3S3NQ29 A0A1B6CPB9 A0A1B6DD07 K7EWQ6 A0A1Y1MRA2 A0A1E1XFV8 A0A3B3T0U0 A0A3Q1EQC9 A0A1A8FCH6 A0A3P8NEQ7 A0A2B4SG71 A0A2B4RPN6 A0A147BCB7 A0A2A4K2Y4 L7MMK9 A0A131XQT5 A0A1B6CK52 A0A1B6E6W0 A0A0S7IPT0 A0A1A8V6A3 W4LLX0 A0A1B6MJ81 A0A3P8VLQ9 A0A0L7L4A5 A0A1B6MUT2 A0A0L7KTF6 X1XU28 A0A3B3H2N7 A0A3P8VSE7 A0A2B4SFB0 L7MC08 A0A3B3H285 A0A2B4RXD0 J9LCT9 A0A2H1V4Y6 A0A3P8Q506 A0A085N6E6 A0A2S2PU23 A0A1B6L0V9 A0A1B6KDA3 A0A2G8KZ95 A0A0L7K286 A0A293MR82 A0A1Z5KXP3 A0A182X639 A0A085LNM9 A0A2B4T0D2 A0A2I4C6V0 A0A3N0YT70 A0A2R5LIR9 A0A0L7K3M9 A0A3B4VPK4 A0A0L7L085 A0A1B8XTH5 A0A3R7GU59 A0A0L7LC80 A0A3B3XP10 A0A1Y1M058 A0A3S0ZV35 A0A1W7R676 A0A1W7R6B8 A0A3B3Q7L2 A2TGR4 A0A085MRV2 W4LLJ5 A0A131YR50 A0A2B4R781 A0A085N4P6
A0A3P8NG55 A0A1Y1LPV1 A0A1A8C840 A0A3S3NQ29 A0A1B6CPB9 A0A1B6DD07 K7EWQ6 A0A1Y1MRA2 A0A1E1XFV8 A0A3B3T0U0 A0A3Q1EQC9 A0A1A8FCH6 A0A3P8NEQ7 A0A2B4SG71 A0A2B4RPN6 A0A147BCB7 A0A2A4K2Y4 L7MMK9 A0A131XQT5 A0A1B6CK52 A0A1B6E6W0 A0A0S7IPT0 A0A1A8V6A3 W4LLX0 A0A1B6MJ81 A0A3P8VLQ9 A0A0L7L4A5 A0A1B6MUT2 A0A0L7KTF6 X1XU28 A0A3B3H2N7 A0A3P8VSE7 A0A2B4SFB0 L7MC08 A0A3B3H285 A0A2B4RXD0 J9LCT9 A0A2H1V4Y6 A0A3P8Q506 A0A085N6E6 A0A2S2PU23 A0A1B6L0V9 A0A1B6KDA3 A0A2G8KZ95 A0A0L7K286 A0A293MR82 A0A1Z5KXP3 A0A182X639 A0A085LNM9 A0A2B4T0D2 A0A2I4C6V0 A0A3N0YT70 A0A2R5LIR9 A0A0L7K3M9 A0A3B4VPK4 A0A0L7L085 A0A1B8XTH5 A0A3R7GU59 A0A0L7LC80 A0A3B3XP10 A0A1Y1M058 A0A3S0ZV35 A0A1W7R676 A0A1W7R6B8 A0A3B3Q7L2 A2TGR4 A0A085MRV2 W4LLJ5 A0A131YR50 A0A2B4R781 A0A085N4P6
Ontologies
KEGG
GO
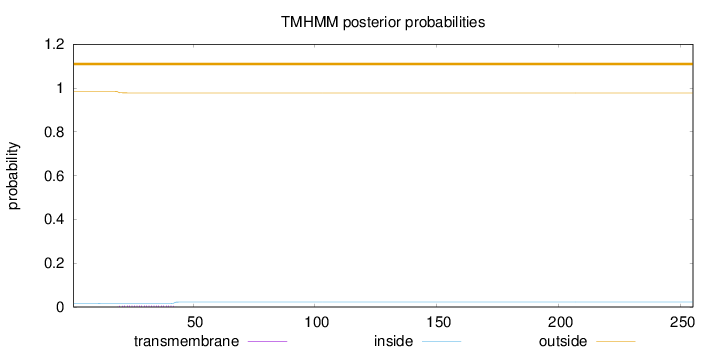
Topology
Length:
255
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.14068
Exp number, first 60 AAs:
0.13925
Total prob of N-in:
0.01657
outside
1 - 255
Population Genetic Test Statistics
Pi
4.625542
Theta
5.122729
Tajima's D
0.234399
CLR
0.585278
CSRT
0.464076796160192
Interpretation
Uncertain
