Gene
KWMTBOMO16431
Pre Gene Modal
BGIBMGA013826
Annotation
PREDICTED:_myb_transcription_factor_isoform_X1_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 4.276
Sequence
CDS
ATGTTCATTGTCAACAGAAAAAGCGGCTATGACAGCGAGACGAGCGAATATTCAGAGGACTCGACGGCGGCTTACGAGGATGTGCAGACTTCAACCAAAAGCACTGGGTCGAGGAAGAATATCAACAGGGGAAGGTGGAGTAAAGAGGAGGACAAGAAACTAAAAGCATATGTTAGGACTTACAAAGAGAATTGGGAGCGCATAGCGGCCGAGTTTCCTGACCGTTCCGACGTTCAGTGCCTTCAAAGATGGACGAAAGTCGTCAACCCTGAACTAGTCAAAGGCCCTTGGACCAAAGAGGAAGACGAGAAAGTCGTTGAGCTGGTCGCGAAATACGGACCGAAGAAATGGACTTTGATAGCCCGACATCTCAAAGGGAGGATAGGGAAGCAGTGCAGAGAGAGATGGCACAATCATCTGAACCCCTGTATAAAGAAGACGGCGTGGACGGAGCACGAGGACAGAGTGATATATCAGGCCCACCAGCAGCTCGGGAACCAGTGGGCGAAGATCGCCAAGCTGTTACCGGGACGAACGGACAACGCAATAAAGAACCATTGGAACTCGACGATGCGTCGTAAATACGACATGGACTTGTTGGACACGCATGAGAGGCGGAAGGGCAAGGCGAAGGCTCGGTTCAACGACGGAGCCCCGGCCGCCGCCCCGGCGGACGAGCGCGTCCAGGTCTCCTACATCGACCAAACCTGGTCGGACCCGTTCAACGAGTCAACCCAGAGCAACGCGTCCAGTACGAGCCAGCCCTCGAAGCAGATGCTCCACTTCCGCCAGGTGCTGAAGAACCAGCTGACGAACCTGCAGCAGCAGCCCCGGCCGGAGTCCCCGGGCCGCGCCGGGCTCGTGGCCGCCGACGCAGAGATCGTTGATTCTCCCTTCAAGTTCGTGAACATCGAGAACCTGCCGACGGACTCGCCGCTCAAGGGCTACCTGAGCCAAGCCCCCGCGGCGGCCGACCCCGGCGGAGACTCCATGTCCTTCTCGATGCACCAGAAGGAGCTGCAGGAGATAGTGGTGCCGGAAGTGTACACGTCGCCGGGCCGGAAGCTGCCGGACGCGAACTCCCCGCCCCCCATCCTGAGGCGGAAGAACAAGAAGCAGATGACGCCTAACCCCAACAACTGGATGGAGTCGATGAGCAAGCTTTCTGAGTTCGGCGCCACCCCGCTGAAGGCGCTGCCGTTCTCCCCCTCGCAGTTCCTCAACTCCCCCCTGGCGCTGCACGCGGCAGACTCCACCCCGCTCCGGGACGCGCACGCCAAGGAGTGGAGCTCGAGCCCGCTGCTGCACACGCCGACGCCGGCCAACAACATCACCCCGGTGCACGTCAACAGGTTCAAGACCAACACTCCCAAAACGCCGACCCCGTTCAAGATCGCGATGGCGGAAATAGGCAAGAAGTCCGGTCTGAAGTACGAGCCGTCCAGCCCCGGACTCCTGGTGGAGGACATCACGGAGATGATCCAGCGGGAGGAGAAGTCGGACAGCACCGTGCAGCTCAGCGACTCCCTGCTCTCCTGCGCCGCCGAGCACGAGGGGGGCGCCCAGGGTAACGACTCCGGCATCGGCAGCCTGAAGCGGCACAGCCTGGACGGGCGGGCGGACGGCAAGGAGAACCTGGCCGCCACGCACAAGCGCGCACGGAAGGCGCTGGCCGCCTCCTGGGGCGCCGCCACCAGCACGCCGCGACACGACGCGCTCGAGACGCCCAGCAAGACCCTGGTCGGGGACAGCTCGGTGCTGTTCTCGCCGCCGTCTATCGTCAAGCACTCTTTGCTGGAGGAGTCCACCAGCCTGCACAGCTACATGAGCGCGGACAGCACGCCCGACGCGCAGCTGGACATCGAGTCCGTCATCAGCGAGACGACCAAGATTGGCGACGACGGCACCTCGGTGACGGTCAAAAGATCCGCCGTCCGGTCAATACGGTTCGAAGACGACCAACCCGACAATGAGTCTGCAGAGGAAATATCCTTTATATTGAGCGACAGTAAGTGGGAGCAGTACGCTTGCGGTAAGACCCAGGATCAAGTGGAATTGACGATGTTAGCCCACCAGTTCCTGAGGAAGACGGCTCTCAAACCGCGCTCGCTCAATTTCTAG
Protein
MFIVNRKSGYDSETSEYSEDSTAAYEDVQTSTKSTGSRKNINRGRWSKEEDKKLKAYVRTYKENWERIAAEFPDRSDVQCLQRWTKVVNPELVKGPWTKEEDEKVVELVAKYGPKKWTLIARHLKGRIGKQCRERWHNHLNPCIKKTAWTEHEDRVIYQAHQQLGNQWAKIAKLLPGRTDNAIKNHWNSTMRRKYDMDLLDTHERRKGKAKARFNDGAPAAAPADERVQVSYIDQTWSDPFNESTQSNASSTSQPSKQMLHFRQVLKNQLTNLQQQPRPESPGRAGLVAADAEIVDSPFKFVNIENLPTDSPLKGYLSQAPAAADPGGDSMSFSMHQKELQEIVVPEVYTSPGRKLPDANSPPPILRRKNKKQMTPNPNNWMESMSKLSEFGATPLKALPFSPSQFLNSPLALHAADSTPLRDAHAKEWSSSPLLHTPTPANNITPVHVNRFKTNTPKTPTPFKIAMAEIGKKSGLKYEPSSPGLLVEDITEMIQREEKSDSTVQLSDSLLSCAAEHEGGAQGNDSGIGSLKRHSLDGRADGKENLAATHKRARKALAASWGAATSTPRHDALETPSKTLVGDSSVLFSPPSIVKHSLLEESTSLHSYMSADSTPDAQLDIESVISETTKIGDDGTSVTVKRSAVRSIRFEDDQPDNESAEEISFILSDSKWEQYACGKTQDQVELTMLAHQFLRKTALKPRSLNF
Summary
Uniprot
A0A3S2LW12
A0A2A4K2N7
A0A2A4K4C8
A0A2A4K432
A0A2A4K2M8
A0A2A4K2M6
+ More
A0A2W1BJM8 A0A2H1VGY9 A0A2A4K4D2 A0A2A4K390 A0A212FK34 A0A194PIH5 A0A2A3EQZ6 A0A0L7QXB3 E2AU28 A0A151JY92 A0A1J1IV61 A0A195BCK3 A0A154NZ50 A0A195DPD5 F4X180 A0A158NFI0 A0A195CV42 A0A151WNW3 A0A3L8E5M5 A0A0M9A517 A0A026WGG2 A0A0P6JNC8 A0A1B6GVG6 A0A2H8THF9 A0A1B6I003 A0A2S2PSS9 A0A0K8SHY4 A0A0A9VZK2 A0A1B6I7M2 A0A0K8T3Y7 A0A0A9VV14 A0A088A0Q8 A0A0K2TVP5 A0A2Y9QW46
A0A2W1BJM8 A0A2H1VGY9 A0A2A4K4D2 A0A2A4K390 A0A212FK34 A0A194PIH5 A0A2A3EQZ6 A0A0L7QXB3 E2AU28 A0A151JY92 A0A1J1IV61 A0A195BCK3 A0A154NZ50 A0A195DPD5 F4X180 A0A158NFI0 A0A195CV42 A0A151WNW3 A0A3L8E5M5 A0A0M9A517 A0A026WGG2 A0A0P6JNC8 A0A1B6GVG6 A0A2H8THF9 A0A1B6I003 A0A2S2PSS9 A0A0K8SHY4 A0A0A9VZK2 A0A1B6I7M2 A0A0K8T3Y7 A0A0A9VV14 A0A088A0Q8 A0A0K2TVP5 A0A2Y9QW46
EMBL
RSAL01000157
RVE45562.1
NWSH01000196
PCG78507.1
PCG78510.1
PCG78513.1
+ More
PCG78505.1 PCG78511.1 KZ150002 PZC75282.1 ODYU01002501 SOQ40077.1 PCG78512.1 PCG78509.1 AGBW02008152 OWR54094.1 KQ459602 KPI93216.1 KZ288201 PBC33559.1 KQ414705 KOC63179.1 GL442767 EFN63071.1 KQ981494 KYN40936.1 CVRI01000061 CRL04063.1 KQ976514 KYM82283.1 KQ434783 KZC04955.1 KQ980662 KYN14706.1 GL888528 EGI59752.1 ADTU01014508 ADTU01014509 ADTU01014510 ADTU01014511 KQ977279 KYN04392.1 KQ982893 KYQ49592.1 QOIP01000001 RLU27605.1 KQ435732 KOX77377.1 KK107260 EZA54154.1 GDIQ01002422 JAN92315.1 GECZ01003394 JAS66375.1 GFXV01001684 MBW13489.1 GECU01027463 JAS80243.1 GGMR01019922 MBY32541.1 GBRD01013097 JAG52729.1 GBHO01045539 GBHO01045537 JAF98064.1 JAF98066.1 GECU01024794 JAS82912.1 GBRD01005926 JAG59895.1 GBHO01045541 GBHO01045536 JAF98062.1 JAF98067.1 HACA01012524 CDW29885.1
PCG78505.1 PCG78511.1 KZ150002 PZC75282.1 ODYU01002501 SOQ40077.1 PCG78512.1 PCG78509.1 AGBW02008152 OWR54094.1 KQ459602 KPI93216.1 KZ288201 PBC33559.1 KQ414705 KOC63179.1 GL442767 EFN63071.1 KQ981494 KYN40936.1 CVRI01000061 CRL04063.1 KQ976514 KYM82283.1 KQ434783 KZC04955.1 KQ980662 KYN14706.1 GL888528 EGI59752.1 ADTU01014508 ADTU01014509 ADTU01014510 ADTU01014511 KQ977279 KYN04392.1 KQ982893 KYQ49592.1 QOIP01000001 RLU27605.1 KQ435732 KOX77377.1 KK107260 EZA54154.1 GDIQ01002422 JAN92315.1 GECZ01003394 JAS66375.1 GFXV01001684 MBW13489.1 GECU01027463 JAS80243.1 GGMR01019922 MBY32541.1 GBRD01013097 JAG52729.1 GBHO01045539 GBHO01045537 JAF98064.1 JAF98066.1 GECU01024794 JAS82912.1 GBRD01005926 JAG59895.1 GBHO01045541 GBHO01045536 JAF98062.1 JAF98067.1 HACA01012524 CDW29885.1
Proteomes
Interpro
SUPFAM
SSF46689
SSF46689
CDD
ProteinModelPortal
A0A3S2LW12
A0A2A4K2N7
A0A2A4K4C8
A0A2A4K432
A0A2A4K2M8
A0A2A4K2M6
+ More
A0A2W1BJM8 A0A2H1VGY9 A0A2A4K4D2 A0A2A4K390 A0A212FK34 A0A194PIH5 A0A2A3EQZ6 A0A0L7QXB3 E2AU28 A0A151JY92 A0A1J1IV61 A0A195BCK3 A0A154NZ50 A0A195DPD5 F4X180 A0A158NFI0 A0A195CV42 A0A151WNW3 A0A3L8E5M5 A0A0M9A517 A0A026WGG2 A0A0P6JNC8 A0A1B6GVG6 A0A2H8THF9 A0A1B6I003 A0A2S2PSS9 A0A0K8SHY4 A0A0A9VZK2 A0A1B6I7M2 A0A0K8T3Y7 A0A0A9VV14 A0A088A0Q8 A0A0K2TVP5 A0A2Y9QW46
A0A2W1BJM8 A0A2H1VGY9 A0A2A4K4D2 A0A2A4K390 A0A212FK34 A0A194PIH5 A0A2A3EQZ6 A0A0L7QXB3 E2AU28 A0A151JY92 A0A1J1IV61 A0A195BCK3 A0A154NZ50 A0A195DPD5 F4X180 A0A158NFI0 A0A195CV42 A0A151WNW3 A0A3L8E5M5 A0A0M9A517 A0A026WGG2 A0A0P6JNC8 A0A1B6GVG6 A0A2H8THF9 A0A1B6I003 A0A2S2PSS9 A0A0K8SHY4 A0A0A9VZK2 A0A1B6I7M2 A0A0K8T3Y7 A0A0A9VV14 A0A088A0Q8 A0A0K2TVP5 A0A2Y9QW46
PDB
1H89
E-value=3.64925e-62,
Score=607
Ontologies
GO
PANTHER
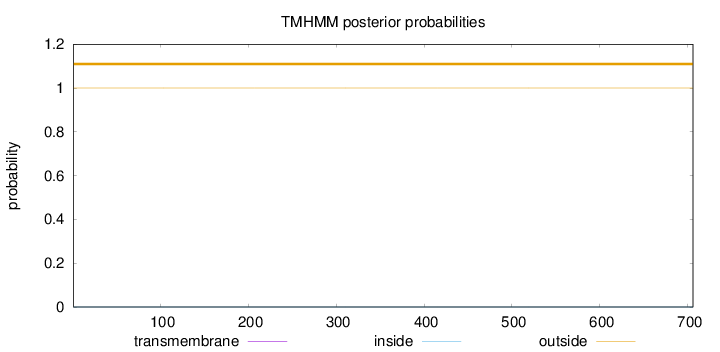
Topology
Subcellular location
Nucleus
Length:
706
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00029
outside
1 - 706
Population Genetic Test Statistics
Pi
250.455602
Theta
169.470144
Tajima's D
1.486872
CLR
0.161646
CSRT
0.786060696965152
Interpretation
Uncertain
