Gene
KWMTBOMO16036
Pre Gene Modal
BGIBMGA004594
Annotation
PREDICTED:_zinc_finger_protein_41-like_isoform_X1_[Amyelois_transitella]
Transcription factor
Location in the cell
Nuclear Reliability : 4.249
Sequence
CDS
ATGGCGTTATTACCCGTTAAGATTGACAAGAACGATCGGCTGCCCCAGAAGATTTGTGATCATTGCAGTCGTAAAGTTAACGAGTTTTATGAGTTTTGTAATGAAACTATTGTAGTGGAAAATAAGCTACAAGCTTTCTTAAAACAGCAAGGAACACACAACACAACTGTAGATCTCATACTTGGAAAACCGAAGACACCAATTCTACCTCCTAAAACATGTGAACAAAGCCATCGAATTGATGAAGGTATACCAGTAGATGAAGTTGTCAAAATTAAAGTTGAAACGCCAAAAGAAATAAAACAAGAAGATATTGAAGCAGAGGAGAACTATCACTACTCAGATGACAGTGATGATGTCTATTTAACAAATCTAAAAGCAAAGAAAAACGATATTGATGTAATAAGATTAAAAAACAGAGAGAAACAGGACGTAAAAAGTAATGGTGTGAAGAAAAATTCATGCGGAAACAAGTCTGAGCTTATGGAAGATTGGTCGATTTTACTAAAGCAGTTCCCTGAGGGCCCTTATAATTTGGTCGACGAAACAGATGTACCAGTTAAACAGGAAGTCAAAGATGAAAACAGTGCAGATTTCGATGACTTGCCTAACTTAAAAGACAAATATCGTTGTTGTATTTGTCTAAAGAATTGCAAGTTTAAGAGTACATTGTTGAAGCACTATAAAACACACGGAAAGGATATTCCAAAGACGAATAATCTTCAGGTGCCTCCAATACCTGATCAGAATCCATTAAAATGCACGAGATGTAGAAAGTCTGTAAAGAAAGATACGTGGGCGGAACATTGGCAGAACCATTGGAAACGCGACCACCGGCCCTACCGATGCGCGTTGTGCGACAAATCGTTCACGTCTTCACAACAAGTCATGGTGCACGGTCTGTCGCACAGCCACAATCCAGTCGACGACGAGGCCATGCAGCTGAAGAAGCAAAATCGTTATGTTTGTGATTTTTGTCCTGAAGAATTTATGTACATGAGATGTCTGCAGGCCCACCGGACGAACGCGCACCCTGAGTCCGCCTCGAAGGCCGTGACGCACCGCTGCTGGAAGTGCTCGCGGCAGTTCGCGCACCTCAATTCCCTGCGGCGGCACTTTATGACGCACACCGGCGAGAGGAACTTCCTGTGCAACGTGTGCGGGAAGGCAATGAGCTCGAGGGACCATCTCAAGCTACACATAAATATACACACCGGCTACAAGCCAAACGTTTGTTCGACATGCGGTAAGGGTTTCGTCAAGAAGAGCAACCTGAAGCTGCACGAGCGCGTCCACAGCGGCGAGAAGCCGTACGTGTGCTCGCACTGCGGGAAGGCGTTCAGCCAGCGATCGACGCTCGTCATACACGAGAGGTGA
Protein
MALLPVKIDKNDRLPQKICDHCSRKVNEFYEFCNETIVVENKLQAFLKQQGTHNTTVDLILGKPKTPILPPKTCEQSHRIDEGIPVDEVVKIKVETPKEIKQEDIEAEENYHYSDDSDDVYLTNLKAKKNDIDVIRLKNREKQDVKSNGVKKNSCGNKSELMEDWSILLKQFPEGPYNLVDETDVPVKQEVKDENSADFDDLPNLKDKYRCCICLKNCKFKSTLLKHYKTHGKDIPKTNNLQVPPIPDQNPLKCTRCRKSVKKDTWAEHWQNHWKRDHRPYRCALCDKSFTSSQQVMVHGLSHSHNPVDDEAMQLKKQNRYVCDFCPEEFMYMRCLQAHRTNAHPESASKAVTHRCWKCSRQFAHLNSLRRHFMTHTGERNFLCNVCGKAMSSRDHLKLHINIHTGYKPNVCSTCGKGFVKKSNLKLHERVHSGEKPYVCSHCGKAFSQRSTLVIHER
Summary
Uniprot
EMBL
BABH01025762
KZ150258
PZC71773.1
NWSH01002922
PCG67292.1
AGBW02008114
+ More
OWR54144.1 KQ459444 KPJ00941.1 KQ460077 KPJ17861.1 ODYU01011531 SOQ57327.1 GEDC01018885 GEDC01018557 GEDC01015550 GEDC01005938 GEDC01004527 JAS18413.1 JAS18741.1 JAS21748.1 JAS31360.1 JAS32771.1 LJIG01001985 KRT84955.1 ODYU01011124 SOQ56669.1 AANG04000264 AHZZ02024315 FR907702 CDQ87145.1
OWR54144.1 KQ459444 KPJ00941.1 KQ460077 KPJ17861.1 ODYU01011531 SOQ57327.1 GEDC01018885 GEDC01018557 GEDC01015550 GEDC01005938 GEDC01004527 JAS18413.1 JAS18741.1 JAS21748.1 JAS31360.1 JAS32771.1 LJIG01001985 KRT84955.1 ODYU01011124 SOQ56669.1 AANG04000264 AHZZ02024315 FR907702 CDQ87145.1
Proteomes
PRIDE
Interpro
ProteinModelPortal
PDB
2I13
E-value=2.62971e-25,
Score=287
Ontologies
GO
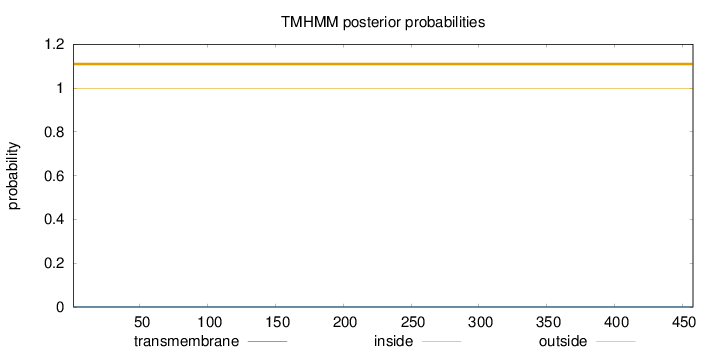
Topology
Length:
458
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00213
outside
1 - 458
Population Genetic Test Statistics
Pi
69.407695
Theta
135.893651
Tajima's D
-0.968187
CLR
146.662085
CSRT
0.143442827858607
Interpretation
Uncertain
