Gene
KWMTBOMO15408
Pre Gene Modal
BGIBMGA004890
Annotation
PREDICTED:_steroid_hormone_receptor_ERR1-like_[Plutella_xylostella]
Transcription factor
Location in the cell
Nuclear Reliability : 3.78
Sequence
CDS
ATGATGTCCGCAGTCAGTGGGGAGCCCATGCTACGCCGCGTCAAACAGGAAACGGATCCGTCGCCGCAGTACTCGCCCCCCGAGCCGCAGCCCCCACACCCCCAGCCGCTGCACCCCCGACCGATGCCCTTGATGCAGCCGGAGGTACCTATGCCTCTCCAACAGTTGGAACTCGAGACTAAGGACGTGAAGTTCTGCGTTTCGCCCGAGGACGGCACGATCGTGGGCAACACCAGCCCTGAGCAGCAGCACTGTTCGTCGACAACCGCGGCCGCCGCCAGCGGCAACCGGGACGAGGACACGCCGCGCAGGCGGTGCCTCGTCTGCGGGGACGTCGCCTCCGGCTTCCACTACGGAGTAGCCAGCTGCGAGGCTTGCAAGGCGTTCTTCAAGAGGACCATACAGGGCAACATCGACTACACTTGTCCGGCGGCGAATGAGTGCGAGATCAACAAGCGGCGGCGCAAGGCGTGCCAGGCGTGCCGCTTCCGCAAGTGCTTGCGCACCGGGATGCTGCGCGAGGGCGTGCGCCTGGACCGCGTGCGCGGCGGCCGCCAGAAGTACCGCCGCGCGCCCGAGCAGGCCCAGCCCAGGGTGCAGCTCGACGACATCAAGGTGCTGGAGGCGCTGCTCTCGTACGAGCCGGAGCCGTTCGCGTGCTCGGCGGCGCCCCCCGGCGTGACGGAGCCGGCCGCGCGGACGCTGGCGCTGCTCGCCGACCTGTACGACCGGGAGCTGGTCGAGATCATCGGCTGGGCCAAGCAGATACCCGGCTTCACGGACCTGCAGCTCAACGATCAGATGCGCTTGTTACAAAGCACTTGGGCCGAGATGCTGAGTCTCATGGTGGCGTACCGCTCGATGTCCAGCGGCGCCAACCTCAAGCTGCGGTTCGCTTCGGACCTCACGCTGGACGAGCAGGAGGCCCGCGACATCGGGGCCACCGATCTGTTTCTACAGGTGTCGGGCGTGGTGCGGCGGCTGGAGCGCGCGGGCGCCGTGCGCGAGGAGTGCGTGCTGCTGAAGGCGCTGGTGCTGGCCAACTCGGAGGCGCGCCTCGACGAGCACCTCACGCTGCGCCGCTTCCGCGAGCAGATCCTGCAGGCGCTCAACGAGGCCGTCGCCGTGCTGCGGTTAGTGACCCACACACACACAGACGTGACGTCATTTGAGGCTATTCGCGCGACGCTATGCTGTCGTGCGTTATGA
Protein
MMSAVSGEPMLRRVKQETDPSPQYSPPEPQPPHPQPLHPRPMPLMQPEVPMPLQQLELETKDVKFCVSPEDGTIVGNTSPEQQHCSSTTAAAASGNRDEDTPRRRCLVCGDVASGFHYGVASCEACKAFFKRTIQGNIDYTCPAANECEINKRRRKACQACRFRKCLRTGMLREGVRLDRVRGGRQKYRRAPEQAQPRVQLDDIKVLEALLSYEPEPFACSAAPPGVTEPAARTLALLADLYDRELVEIIGWAKQIPGFTDLQLNDQMRLLQSTWAEMLSLMVAYRSMSSGANLKLRFASDLTLDEQEARDIGATDLFLQVSGVVRRLERAGAVREECVLLKALVLANSEARLDEHLTLRRFREQILQALNEAVAVLRLVTHTHTDVTSFEAIRATLCCRAL
Summary
Similarity
Belongs to the nuclear hormone receptor family.
Belongs to the nuclear hormone receptor family. NR3 subfamily.
Belongs to the nuclear hormone receptor family. NR3 subfamily.
Uniprot
A0A1B1M0R7
A0A2A4JYX1
A0A2A4K0Q3
A0A2H1V335
A0A1I9RYC3
A0A3S2NSI7
+ More
A0A2W1BIB8 A0A212FQ16 A0A0N0PBA0 A0A194QDI5 A0A0U2S7G3 A0A0L7LNK9 D6WU20 A0A139WFK8 A0A1Y1L3V2 A0A1Y1L2Q4 A0A2J7QLP3 U4U3T1 A0A1W4X7V6 N6UKY6 A0A1W4WYG7 A0A067QT68 A0A2P8YW51 A0A1S5UZQ5 A0A223HFU7 A0A1D9C6F9 A0A221C9P7 A0A346TJJ9 A0A1B6JZQ2 D2Y1A7 A0A1B6JJE4 A0A097C257 E0VYP3 F4WPT4 A0A226EV34 D1LUZ3 A0A1B6MIK9 A0A1B6MNL6 E2BSJ6 A0A0A9VSR2 C1I1I1 A0A195EH28 A0A1S4FZE4 A0A1J1ITD3 Q16IU3 A0A0J7KHQ3 A0A195CKQ9 A0A0A9Y8J4 A0A151WJS5 A0A0V0GD90 A0A1D2NKF5 A0A0C9RQH6 A0A069DUC1 A0A158NI98 A0A1I9J8G2 C4MBZ4 A0A195AX26 K7J6M1 U5EWZ8 A0A026WWP8 D8L549 B0WYX7 A0A0L7R722 A0A195F662 A0A088A026 A0A131XRQ5 A0A0N0BJF8 A0A1Q3FYJ5 A0A1Q3FYV5 A0A1Q3FYM6 A0A224XJD6 A0A0A9YA46 A0A087U0Z5 A0A0A9VQS0 A0A1B0GMR7 A0A0A7CJW0 A0A0A7CK37 T1IPY4 A0A1L8DH32 A0A1Z5L1L1 A0A1L8DKI4 E2B1G4 A0A336KLY2 A0A1E1XJQ8 A0A2R5LLU9 E3W3U3 A0A0P5C7Q2 A0A0P5C7F6 A0A131YGL7 L7LXI7 A0A1E1X399 A0A0P5C7H7 A0A0P6ASE0 A0A0P5GAA9 A0A0N8EGQ2 A0A0P5SRZ1 A0A0P6BCV6 T1GCB1 A0A0P5RRU2
A0A2W1BIB8 A0A212FQ16 A0A0N0PBA0 A0A194QDI5 A0A0U2S7G3 A0A0L7LNK9 D6WU20 A0A139WFK8 A0A1Y1L3V2 A0A1Y1L2Q4 A0A2J7QLP3 U4U3T1 A0A1W4X7V6 N6UKY6 A0A1W4WYG7 A0A067QT68 A0A2P8YW51 A0A1S5UZQ5 A0A223HFU7 A0A1D9C6F9 A0A221C9P7 A0A346TJJ9 A0A1B6JZQ2 D2Y1A7 A0A1B6JJE4 A0A097C257 E0VYP3 F4WPT4 A0A226EV34 D1LUZ3 A0A1B6MIK9 A0A1B6MNL6 E2BSJ6 A0A0A9VSR2 C1I1I1 A0A195EH28 A0A1S4FZE4 A0A1J1ITD3 Q16IU3 A0A0J7KHQ3 A0A195CKQ9 A0A0A9Y8J4 A0A151WJS5 A0A0V0GD90 A0A1D2NKF5 A0A0C9RQH6 A0A069DUC1 A0A158NI98 A0A1I9J8G2 C4MBZ4 A0A195AX26 K7J6M1 U5EWZ8 A0A026WWP8 D8L549 B0WYX7 A0A0L7R722 A0A195F662 A0A088A026 A0A131XRQ5 A0A0N0BJF8 A0A1Q3FYJ5 A0A1Q3FYV5 A0A1Q3FYM6 A0A224XJD6 A0A0A9YA46 A0A087U0Z5 A0A0A9VQS0 A0A1B0GMR7 A0A0A7CJW0 A0A0A7CK37 T1IPY4 A0A1L8DH32 A0A1Z5L1L1 A0A1L8DKI4 E2B1G4 A0A336KLY2 A0A1E1XJQ8 A0A2R5LLU9 E3W3U3 A0A0P5C7Q2 A0A0P5C7F6 A0A131YGL7 L7LXI7 A0A1E1X399 A0A0P5C7H7 A0A0P6ASE0 A0A0P5GAA9 A0A0N8EGQ2 A0A0P5SRZ1 A0A0P6BCV6 T1GCB1 A0A0P5RRU2
Pubmed
EMBL
KT268294
ANS60466.1
NWSH01000346
PCG77221.1
PCG77222.1
ODYU01000450
+ More
SOQ35255.1 KT944662 AOZ56905.1 RSAL01000001 RVE55258.1 KZ150138 PZC72967.1 AGBW02000323 OWR55793.1 KQ460973 KPJ10088.1 KQ459324 KPJ01521.1 KU301327 ALT07179.1 JTDY01000450 KOB77123.1 KQ971352 EFA07462.2 KYB26704.1 GEZM01068290 JAV67040.1 GEZM01068289 JAV67041.1 NEVH01013243 PNF29496.1 KB631985 ERL87717.1 APGK01019570 KB740112 ENN81341.1 KK853567 KDR06670.1 PYGN01000325 PSN48477.1 KX373870 AQN67829.1 KX431895 AST48089.1 KU899089 AOY10609.1 KY210878 ASL70079.1 MF537045 AXU37748.1 GECU01003034 JAT04673.1 GU290313 ADB43256.1 GECU01008401 JAS99305.1 KM189915 AIS76179.1 DS235845 EEB18499.1 GL888255 EGI63763.1 LNIX01000002 OXA60934.1 GU070740 ACY66850.1 GEBQ01004211 JAT35766.1 GEBQ01002518 JAT37459.1 GL450240 EFN81329.1 GBHO01044835 GBHO01021855 GBHO01021854 GBRD01009378 GDHC01006753 JAF98768.1 JAG21749.1 JAG21750.1 JAG56446.1 JAQ11876.1 EF474463 ABR88112.1 KQ978957 KYN27174.1 CVRI01000059 CRL02986.1 CH478052 EAT34188.1 LBMM01007381 KMQ89782.1 KQ977622 KYN01293.1 GBHO01015126 GBHO01015125 GBHO01015124 GDHC01021143 JAG28478.1 JAG28479.1 JAG28480.1 JAP97485.1 KQ983039 KYQ48084.1 GECL01000227 JAP05897.1 LJIJ01000018 ODN05709.1 GBYB01009471 GBYB01009472 JAG79238.1 JAG79239.1 GBGD01001582 JAC87307.1 ADTU01016132 ADTU01016133 ADTU01016134 ADTU01016135 KP398511 AKQ06251.1 EF506198 ABS50436.1 KQ976725 KYM76514.1 AAZX01007930 GANO01000318 JAB59553.1 KK107078 EZA60253.1 FJ770332 ACW84414.1 DS232200 EDS37237.1 KQ414643 KOC66682.1 KQ981763 KYN36080.1 GEFM01005823 GEGO01002978 JAP69973.1 JAR92426.1 KQ435719 KOX78864.1 GFDL01002401 JAV32644.1 GFDL01002393 JAV32652.1 GFDL01002394 JAV32651.1 GFTR01005262 JAW11164.1 GBHO01015128 GBHO01015118 GBHO01015117 JAG28476.1 JAG28486.1 JAG28487.1 KK117641 KFM71034.1 GBHO01044822 GBHO01021856 GBHO01015122 JAF98781.1 JAG21748.1 JAG28482.1 AJVK01026433 KJ664215 AID52852.1 KJ664216 AID52853.1 JH431265 GFDF01008353 JAV05731.1 GFJQ02006006 JAW00964.1 GFDF01007118 JAV06966.1 GL444918 EFN60472.1 UFQS01000437 UFQT01000437 SSX03933.1 SSX24298.1 GFAA01003956 JAT99478.1 GGLE01006282 MBY10408.1 HQ386702 ADO51071.1 GDIP01174371 JAJ49031.1 GDIP01174372 JAJ49030.1 GEDV01010123 JAP78434.1 GACK01008732 JAA56302.1 GFAC01005461 JAT93727.1 GDIP01174370 JAJ49032.1 GDIP01039058 LRGB01000337 JAM64657.1 KZS19653.1 GDIQ01245247 JAK06478.1 GDIQ01028452 JAN66285.1 GDIP01136170 JAL67544.1 GDIP01017011 JAM86704.1 CAQQ02186414 CAQQ02186415 GDIQ01102741 JAL48985.1
SOQ35255.1 KT944662 AOZ56905.1 RSAL01000001 RVE55258.1 KZ150138 PZC72967.1 AGBW02000323 OWR55793.1 KQ460973 KPJ10088.1 KQ459324 KPJ01521.1 KU301327 ALT07179.1 JTDY01000450 KOB77123.1 KQ971352 EFA07462.2 KYB26704.1 GEZM01068290 JAV67040.1 GEZM01068289 JAV67041.1 NEVH01013243 PNF29496.1 KB631985 ERL87717.1 APGK01019570 KB740112 ENN81341.1 KK853567 KDR06670.1 PYGN01000325 PSN48477.1 KX373870 AQN67829.1 KX431895 AST48089.1 KU899089 AOY10609.1 KY210878 ASL70079.1 MF537045 AXU37748.1 GECU01003034 JAT04673.1 GU290313 ADB43256.1 GECU01008401 JAS99305.1 KM189915 AIS76179.1 DS235845 EEB18499.1 GL888255 EGI63763.1 LNIX01000002 OXA60934.1 GU070740 ACY66850.1 GEBQ01004211 JAT35766.1 GEBQ01002518 JAT37459.1 GL450240 EFN81329.1 GBHO01044835 GBHO01021855 GBHO01021854 GBRD01009378 GDHC01006753 JAF98768.1 JAG21749.1 JAG21750.1 JAG56446.1 JAQ11876.1 EF474463 ABR88112.1 KQ978957 KYN27174.1 CVRI01000059 CRL02986.1 CH478052 EAT34188.1 LBMM01007381 KMQ89782.1 KQ977622 KYN01293.1 GBHO01015126 GBHO01015125 GBHO01015124 GDHC01021143 JAG28478.1 JAG28479.1 JAG28480.1 JAP97485.1 KQ983039 KYQ48084.1 GECL01000227 JAP05897.1 LJIJ01000018 ODN05709.1 GBYB01009471 GBYB01009472 JAG79238.1 JAG79239.1 GBGD01001582 JAC87307.1 ADTU01016132 ADTU01016133 ADTU01016134 ADTU01016135 KP398511 AKQ06251.1 EF506198 ABS50436.1 KQ976725 KYM76514.1 AAZX01007930 GANO01000318 JAB59553.1 KK107078 EZA60253.1 FJ770332 ACW84414.1 DS232200 EDS37237.1 KQ414643 KOC66682.1 KQ981763 KYN36080.1 GEFM01005823 GEGO01002978 JAP69973.1 JAR92426.1 KQ435719 KOX78864.1 GFDL01002401 JAV32644.1 GFDL01002393 JAV32652.1 GFDL01002394 JAV32651.1 GFTR01005262 JAW11164.1 GBHO01015128 GBHO01015118 GBHO01015117 JAG28476.1 JAG28486.1 JAG28487.1 KK117641 KFM71034.1 GBHO01044822 GBHO01021856 GBHO01015122 JAF98781.1 JAG21748.1 JAG28482.1 AJVK01026433 KJ664215 AID52852.1 KJ664216 AID52853.1 JH431265 GFDF01008353 JAV05731.1 GFJQ02006006 JAW00964.1 GFDF01007118 JAV06966.1 GL444918 EFN60472.1 UFQS01000437 UFQT01000437 SSX03933.1 SSX24298.1 GFAA01003956 JAT99478.1 GGLE01006282 MBY10408.1 HQ386702 ADO51071.1 GDIP01174371 JAJ49031.1 GDIP01174372 JAJ49030.1 GEDV01010123 JAP78434.1 GACK01008732 JAA56302.1 GFAC01005461 JAT93727.1 GDIP01174370 JAJ49032.1 GDIP01039058 LRGB01000337 JAM64657.1 KZS19653.1 GDIQ01245247 JAK06478.1 GDIQ01028452 JAN66285.1 GDIP01136170 JAL67544.1 GDIP01017011 JAM86704.1 CAQQ02186414 CAQQ02186415 GDIQ01102741 JAL48985.1
Proteomes
UP000218220
UP000283053
UP000007151
UP000053240
UP000053268
UP000037510
+ More
UP000007266 UP000235965 UP000030742 UP000192223 UP000019118 UP000027135 UP000245037 UP000009046 UP000007755 UP000198287 UP000008237 UP000078492 UP000183832 UP000008820 UP000036403 UP000078542 UP000075809 UP000094527 UP000005205 UP000078540 UP000002358 UP000053097 UP000002320 UP000053825 UP000078541 UP000005203 UP000053105 UP000054359 UP000092462 UP000000311 UP000076858 UP000015102
UP000007266 UP000235965 UP000030742 UP000192223 UP000019118 UP000027135 UP000245037 UP000009046 UP000007755 UP000198287 UP000008237 UP000078492 UP000183832 UP000008820 UP000036403 UP000078542 UP000075809 UP000094527 UP000005205 UP000078540 UP000002358 UP000053097 UP000002320 UP000053825 UP000078541 UP000005203 UP000053105 UP000054359 UP000092462 UP000000311 UP000076858 UP000015102
Interpro
SUPFAM
SSF48508
SSF48508
Gene 3D
ProteinModelPortal
A0A1B1M0R7
A0A2A4JYX1
A0A2A4K0Q3
A0A2H1V335
A0A1I9RYC3
A0A3S2NSI7
+ More
A0A2W1BIB8 A0A212FQ16 A0A0N0PBA0 A0A194QDI5 A0A0U2S7G3 A0A0L7LNK9 D6WU20 A0A139WFK8 A0A1Y1L3V2 A0A1Y1L2Q4 A0A2J7QLP3 U4U3T1 A0A1W4X7V6 N6UKY6 A0A1W4WYG7 A0A067QT68 A0A2P8YW51 A0A1S5UZQ5 A0A223HFU7 A0A1D9C6F9 A0A221C9P7 A0A346TJJ9 A0A1B6JZQ2 D2Y1A7 A0A1B6JJE4 A0A097C257 E0VYP3 F4WPT4 A0A226EV34 D1LUZ3 A0A1B6MIK9 A0A1B6MNL6 E2BSJ6 A0A0A9VSR2 C1I1I1 A0A195EH28 A0A1S4FZE4 A0A1J1ITD3 Q16IU3 A0A0J7KHQ3 A0A195CKQ9 A0A0A9Y8J4 A0A151WJS5 A0A0V0GD90 A0A1D2NKF5 A0A0C9RQH6 A0A069DUC1 A0A158NI98 A0A1I9J8G2 C4MBZ4 A0A195AX26 K7J6M1 U5EWZ8 A0A026WWP8 D8L549 B0WYX7 A0A0L7R722 A0A195F662 A0A088A026 A0A131XRQ5 A0A0N0BJF8 A0A1Q3FYJ5 A0A1Q3FYV5 A0A1Q3FYM6 A0A224XJD6 A0A0A9YA46 A0A087U0Z5 A0A0A9VQS0 A0A1B0GMR7 A0A0A7CJW0 A0A0A7CK37 T1IPY4 A0A1L8DH32 A0A1Z5L1L1 A0A1L8DKI4 E2B1G4 A0A336KLY2 A0A1E1XJQ8 A0A2R5LLU9 E3W3U3 A0A0P5C7Q2 A0A0P5C7F6 A0A131YGL7 L7LXI7 A0A1E1X399 A0A0P5C7H7 A0A0P6ASE0 A0A0P5GAA9 A0A0N8EGQ2 A0A0P5SRZ1 A0A0P6BCV6 T1GCB1 A0A0P5RRU2
A0A2W1BIB8 A0A212FQ16 A0A0N0PBA0 A0A194QDI5 A0A0U2S7G3 A0A0L7LNK9 D6WU20 A0A139WFK8 A0A1Y1L3V2 A0A1Y1L2Q4 A0A2J7QLP3 U4U3T1 A0A1W4X7V6 N6UKY6 A0A1W4WYG7 A0A067QT68 A0A2P8YW51 A0A1S5UZQ5 A0A223HFU7 A0A1D9C6F9 A0A221C9P7 A0A346TJJ9 A0A1B6JZQ2 D2Y1A7 A0A1B6JJE4 A0A097C257 E0VYP3 F4WPT4 A0A226EV34 D1LUZ3 A0A1B6MIK9 A0A1B6MNL6 E2BSJ6 A0A0A9VSR2 C1I1I1 A0A195EH28 A0A1S4FZE4 A0A1J1ITD3 Q16IU3 A0A0J7KHQ3 A0A195CKQ9 A0A0A9Y8J4 A0A151WJS5 A0A0V0GD90 A0A1D2NKF5 A0A0C9RQH6 A0A069DUC1 A0A158NI98 A0A1I9J8G2 C4MBZ4 A0A195AX26 K7J6M1 U5EWZ8 A0A026WWP8 D8L549 B0WYX7 A0A0L7R722 A0A195F662 A0A088A026 A0A131XRQ5 A0A0N0BJF8 A0A1Q3FYJ5 A0A1Q3FYV5 A0A1Q3FYM6 A0A224XJD6 A0A0A9YA46 A0A087U0Z5 A0A0A9VQS0 A0A1B0GMR7 A0A0A7CJW0 A0A0A7CK37 T1IPY4 A0A1L8DH32 A0A1Z5L1L1 A0A1L8DKI4 E2B1G4 A0A336KLY2 A0A1E1XJQ8 A0A2R5LLU9 E3W3U3 A0A0P5C7Q2 A0A0P5C7F6 A0A131YGL7 L7LXI7 A0A1E1X399 A0A0P5C7H7 A0A0P6ASE0 A0A0P5GAA9 A0A0N8EGQ2 A0A0P5SRZ1 A0A0P6BCV6 T1GCB1 A0A0P5RRU2
PDB
1LO1
E-value=9.55176e-40,
Score=411
Ontologies
GO
Topology
Subcellular location
Nucleus
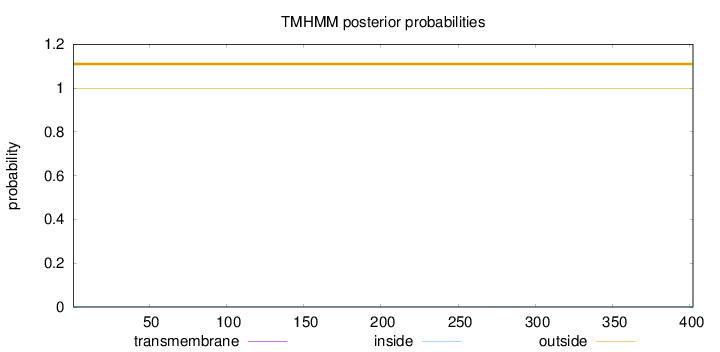
Length:
402
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00571999999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00300
outside
1 - 402
Population Genetic Test Statistics
Pi
196.785256
Theta
167.592214
Tajima's D
0.544447
CLR
0.446909
CSRT
0.525823708814559
Interpretation
Uncertain
