Gene
KWMTBOMO15162
Pre Gene Modal
BGIBMGA004983
Annotation
Lipopolysaccharide-induced_tumor_necrosis_factor-alpha_factor-like_protein_[Operophtera_brumata]
Full name
Lipopolysaccharide-induced tumor necrosis factor-alpha factor homolog
Alternative Name
Small integral membrane protein of lysosome/late endosome
Estrogen-enhanced transcript protein 1
Estrogen-enhanced transcript protein 1
Transcription factor
Location in the cell
Extracellular Reliability : 2.662
Sequence
CDS
ATGTCTACCCATCAAGGACCTAGTGCGCCACTAGACGATCTTCCACCGCCGTACTCATCGGTTGTAGGAACTACGCATTATGGCTTTGTGGCTCCGGCACTAAGTAATCCTATCCCGGATTCTGTAGGAGCATATCCACCTGCTAAACCATTTACTCCTCCAGGAGTTTACCCCCATCCCGAAGGTGTAACAAATGCAACTGTAAATATGCAGCCTTCTAGAATACCACCTCCACCACCAGGCTTACCAGTCGGTATTGTTCTTCCAAACGCTATGGGTACAGAACCAACCACTTTAACATGTTTCAACTGTAATAAAATAGTTACTACAAGAGTTACATACACAACAGCTTGGCACACACATTTAATTGCTGGGTCGATTTGCTTGATCACTATAGTTTGTTCACTATGCTGCCTGGGTTTGATCCCGTACTGCTTTGATACATTCAAGGAGGCTGAGCACTACTGCCCTAATTGCAACACATTCATCGGCAAAAGCAACAAGTGTTAA
Protein
MSTHQGPSAPLDDLPPPYSSVVGTTHYGFVAPALSNPIPDSVGAYPPAKPFTPPGVYPHPEGVTNATVNMQPSRIPPPPPGLPVGIVLPNAMGTEPTTLTCFNCNKIVTTRVTYTTAWHTHLIAGSICLITIVCSLCCLGLIPYCFDTFKEAEHYCPNCNTFIGKSNKC
Summary
Description
Plays a role in endosomal protein trafficking and in targeting proteins for lysosomal degradation. May also contribute to the regulation of gene expression in the nucleus. Binds DNA (in vitro) and may play a synergistic role in the nucleus in regulating the expression of numerous cytokines.
Plays a role in endosomal protein trafficking and in targeting proteins for lysosomal degradation. Plays a role in targeting endocytosed EGFR and ERGG3 for lysosomal degradation, and thereby helps downregulate downstream signaling cascades. Helps recruit the ESCRT complex components TSG101, HGS and STAM to cytoplasmic membranes. Probably plays a role in regulating protein degradation via its interaction with NEDD4. May also contribute to the regulation of gene expression in the nucleus. Binds DNA (in vitro) and may play a synergistic role with STAT6 in the nucleus in regulating the expression of various cytokines. May regulate the expression of numerous cytokines, such as TNF, CCL2, CCL5, CXCL1, IL1A and IL10.
Plays a role in endosomal protein trafficking and in targeting proteins for lysosomal degradation. Plays a role in targeting endocytosed EGFR and ERGG3 for lysosomal degradation, and thereby helps downregulate downstream signaling cascades. Helps recruit the ESCRT complex components TSG101, HGS and STAM to cytoplasmic membranes. Probably plays a role in regulating protein degradation via its interaction with NEDD4. May also contribute to the regulation of gene expression in the nucleus. Binds DNA (in vitro) and may play a synergistic role with STAT6 in the nucleus in regulating the expression of various cytokines. May regulate the expression of numerous cytokines, such as TNF, CCL2, CCL5, CXCL1, IL1A and IL10.
Subunit
Monomer. Interacts with NEDD4. Interacts (via PSAP motif) with TSG101, a component of the ESCRT-I complex (endosomal sorting complex required for transport I). Interacts with WWOX. Interacts with STAM, a component of the ESCRT-0 complex; the interaction is direct. Identified in a complex with STAM and HGS; within this complex, interacts directly with STAM, but not with HGS. Interacts with STAT6.
Similarity
Belongs to the CDIP1/LITAF family.
Keywords
Cell membrane
Complete proteome
Cytoplasm
DNA-binding
Endosome
Golgi apparatus
Lysosome
Membrane
Metal-binding
Nucleus
Reference proteome
Transcription
Transcription regulation
Zinc
Feature
chain Lipopolysaccharide-induced tumor necrosis factor-alpha factor homolog
Uniprot
H9J642
A0A0L7LPB8
A0A2H1WQ02
A0A194RKY7
A0A194PME3
S4PR50
+ More
A0A212EZ24 G5B4G8 A0A336MCY4 G3WH57 A0A0N8ET20 L5KQM0 S7PRL7 A0A340XRQ9 A0A2P4TB50 A0A3Q2LLF6 A0A093K5J9 F6PP17 A0A3Q2LDN3 H0V8A5 A0A091U0I7 A0A3Q2H8I3 A0A093DXW7 F1NNL3 A0A2Y9SRV5 A0A2Y9QL88 A0A091CRW7 A0A341BE27 A0A2Y9LLU0 G3STV5 Q8QGW7 A0A2Y9QXY7 A0A2Y9LGY8 A0A091H2N9 S9XMI4 B2RYP2 P0C0T0 A0A091FZU0 A0A2Y9HPR8 A0A2U3Z2R2 A0A1A6GSL7 A0A2U3W0C3 A0A1U7QZX7 G3GWL3 K9IYC9
A0A212EZ24 G5B4G8 A0A336MCY4 G3WH57 A0A0N8ET20 L5KQM0 S7PRL7 A0A340XRQ9 A0A2P4TB50 A0A3Q2LLF6 A0A093K5J9 F6PP17 A0A3Q2LDN3 H0V8A5 A0A091U0I7 A0A3Q2H8I3 A0A093DXW7 F1NNL3 A0A2Y9SRV5 A0A2Y9QL88 A0A091CRW7 A0A341BE27 A0A2Y9LLU0 G3STV5 Q8QGW7 A0A2Y9QXY7 A0A2Y9LGY8 A0A091H2N9 S9XMI4 B2RYP2 P0C0T0 A0A091FZU0 A0A2Y9HPR8 A0A2U3Z2R2 A0A1A6GSL7 A0A2U3W0C3 A0A1U7QZX7 G3GWL3 K9IYC9
Pubmed
EMBL
BABH01029946
JTDY01000430
KOB77214.1
ODYU01010192
SOQ55151.1
KQ460045
+ More
KPJ18207.1 KQ459604 KPI92285.1 GAIX01001120 JAA91440.1 AGBW02011379 OWR46749.1 JH168449 EHB04179.1 UFQT01000784 SSX27211.1 AEFK01077910 GEBF01005603 JAN98029.1 KB030624 ELK13525.1 KE163197 EPQ11147.1 PPHD01003233 POI33592.1 KL206788 KFV85729.1 AAKN02007517 KK408314 KFQ83365.1 KL232961 KFV07011.1 AADN05000376 KN124499 KFO20992.1 AB058634 AB061737 AY765397 KL518851 KFO88930.1 KB017616 EPY76391.1 BC166851 CH474017 AAI66851.1 EDL96205.1 U53184 KL447645 KFO75750.1 LZPO01075857 OBS68899.1 JH000052 RAZU01000241 EGV96323.1 RLQ63293.1 GABZ01008306 JAA45219.1
KPJ18207.1 KQ459604 KPI92285.1 GAIX01001120 JAA91440.1 AGBW02011379 OWR46749.1 JH168449 EHB04179.1 UFQT01000784 SSX27211.1 AEFK01077910 GEBF01005603 JAN98029.1 KB030624 ELK13525.1 KE163197 EPQ11147.1 PPHD01003233 POI33592.1 KL206788 KFV85729.1 AAKN02007517 KK408314 KFQ83365.1 KL232961 KFV07011.1 AADN05000376 KN124499 KFO20992.1 AB058634 AB061737 AY765397 KL518851 KFO88930.1 KB017616 EPY76391.1 BC166851 CH474017 AAI66851.1 EDL96205.1 U53184 KL447645 KFO75750.1 LZPO01075857 OBS68899.1 JH000052 RAZU01000241 EGV96323.1 RLQ63293.1 GABZ01008306 JAA45219.1
Proteomes
UP000005204
UP000037510
UP000053240
UP000053268
UP000007151
UP000006813
+ More
UP000007648 UP000010552 UP000265300 UP000002281 UP000053584 UP000005447 UP000000539 UP000248484 UP000248480 UP000028990 UP000252040 UP000248483 UP000007646 UP000002494 UP000053760 UP000248481 UP000245341 UP000092124 UP000245340 UP000189706 UP000001075 UP000273346
UP000007648 UP000010552 UP000265300 UP000002281 UP000053584 UP000005447 UP000000539 UP000248484 UP000248480 UP000028990 UP000252040 UP000248483 UP000007646 UP000002494 UP000053760 UP000248481 UP000245341 UP000092124 UP000245340 UP000189706 UP000001075 UP000273346
PRIDE
Pfam
PF10601 zf-LITAF-like
ProteinModelPortal
H9J642
A0A0L7LPB8
A0A2H1WQ02
A0A194RKY7
A0A194PME3
S4PR50
+ More
A0A212EZ24 G5B4G8 A0A336MCY4 G3WH57 A0A0N8ET20 L5KQM0 S7PRL7 A0A340XRQ9 A0A2P4TB50 A0A3Q2LLF6 A0A093K5J9 F6PP17 A0A3Q2LDN3 H0V8A5 A0A091U0I7 A0A3Q2H8I3 A0A093DXW7 F1NNL3 A0A2Y9SRV5 A0A2Y9QL88 A0A091CRW7 A0A341BE27 A0A2Y9LLU0 G3STV5 Q8QGW7 A0A2Y9QXY7 A0A2Y9LGY8 A0A091H2N9 S9XMI4 B2RYP2 P0C0T0 A0A091FZU0 A0A2Y9HPR8 A0A2U3Z2R2 A0A1A6GSL7 A0A2U3W0C3 A0A1U7QZX7 G3GWL3 K9IYC9
A0A212EZ24 G5B4G8 A0A336MCY4 G3WH57 A0A0N8ET20 L5KQM0 S7PRL7 A0A340XRQ9 A0A2P4TB50 A0A3Q2LLF6 A0A093K5J9 F6PP17 A0A3Q2LDN3 H0V8A5 A0A091U0I7 A0A3Q2H8I3 A0A093DXW7 F1NNL3 A0A2Y9SRV5 A0A2Y9QL88 A0A091CRW7 A0A341BE27 A0A2Y9LLU0 G3STV5 Q8QGW7 A0A2Y9QXY7 A0A2Y9LGY8 A0A091H2N9 S9XMI4 B2RYP2 P0C0T0 A0A091FZU0 A0A2Y9HPR8 A0A2U3Z2R2 A0A1A6GSL7 A0A2U3W0C3 A0A1U7QZX7 G3GWL3 K9IYC9
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasm
Associated with membranes of lysosomes, early and late endosomes. Can translocate from the cytoplasm into the nucleus (By similarity). Detected at Schmidt-Lanterman incisures and in nodal regions of myelinating Schwann cells (By similarity). With evidence from 1 publications.
Nucleus Associated with membranes of lysosomes, early and late endosomes. Can translocate from the cytoplasm into the nucleus (By similarity). Detected at Schmidt-Lanterman incisures and in nodal regions of myelinating Schwann cells (By similarity). With evidence from 1 publications.
Lysosome membrane Associated with membranes of lysosomes, early and late endosomes. Can translocate from the cytoplasm into the nucleus (By similarity). Detected at Schmidt-Lanterman incisures and in nodal regions of myelinating Schwann cells (By similarity). With evidence from 1 publications.
Early endosome membrane Associated with membranes of lysosomes, early and late endosomes. Can translocate from the cytoplasm into the nucleus (By similarity). Detected at Schmidt-Lanterman incisures and in nodal regions of myelinating Schwann cells (By similarity). With evidence from 1 publications.
Late endosome membrane Associated with membranes of lysosomes, early and late endosomes. Can translocate from the cytoplasm into the nucleus (By similarity). Detected at Schmidt-Lanterman incisures and in nodal regions of myelinating Schwann cells (By similarity). With evidence from 1 publications.
Endosome membrane Associated with membranes of lysosomes, early and late endosomes. Can translocate from the cytoplasm into the nucleus (By similarity). Detected at Schmidt-Lanterman incisures and in nodal regions of myelinating Schwann cells (By similarity). With evidence from 1 publications.
Cell membrane Associated with membranes of lysosomes, early and late endosomes. Can translocate from the cytoplasm into the nucleus (By similarity). Detected at Schmidt-Lanterman incisures and in nodal regions of myelinating Schwann cells (By similarity). With evidence from 1 publications.
Golgi apparatus membrane Associated with membranes of lysosomes, early and late endosomes. Can translocate from the cytoplasm into the nucleus (By similarity). Detected at Schmidt-Lanterman incisures and in nodal regions of myelinating Schwann cells (By similarity). With evidence from 1 publications.
Nucleus Associated with membranes of lysosomes, early and late endosomes. Can translocate from the cytoplasm into the nucleus (By similarity). Detected at Schmidt-Lanterman incisures and in nodal regions of myelinating Schwann cells (By similarity). With evidence from 1 publications.
Lysosome membrane Associated with membranes of lysosomes, early and late endosomes. Can translocate from the cytoplasm into the nucleus (By similarity). Detected at Schmidt-Lanterman incisures and in nodal regions of myelinating Schwann cells (By similarity). With evidence from 1 publications.
Early endosome membrane Associated with membranes of lysosomes, early and late endosomes. Can translocate from the cytoplasm into the nucleus (By similarity). Detected at Schmidt-Lanterman incisures and in nodal regions of myelinating Schwann cells (By similarity). With evidence from 1 publications.
Late endosome membrane Associated with membranes of lysosomes, early and late endosomes. Can translocate from the cytoplasm into the nucleus (By similarity). Detected at Schmidt-Lanterman incisures and in nodal regions of myelinating Schwann cells (By similarity). With evidence from 1 publications.
Endosome membrane Associated with membranes of lysosomes, early and late endosomes. Can translocate from the cytoplasm into the nucleus (By similarity). Detected at Schmidt-Lanterman incisures and in nodal regions of myelinating Schwann cells (By similarity). With evidence from 1 publications.
Cell membrane Associated with membranes of lysosomes, early and late endosomes. Can translocate from the cytoplasm into the nucleus (By similarity). Detected at Schmidt-Lanterman incisures and in nodal regions of myelinating Schwann cells (By similarity). With evidence from 1 publications.
Golgi apparatus membrane Associated with membranes of lysosomes, early and late endosomes. Can translocate from the cytoplasm into the nucleus (By similarity). Detected at Schmidt-Lanterman incisures and in nodal regions of myelinating Schwann cells (By similarity). With evidence from 1 publications.
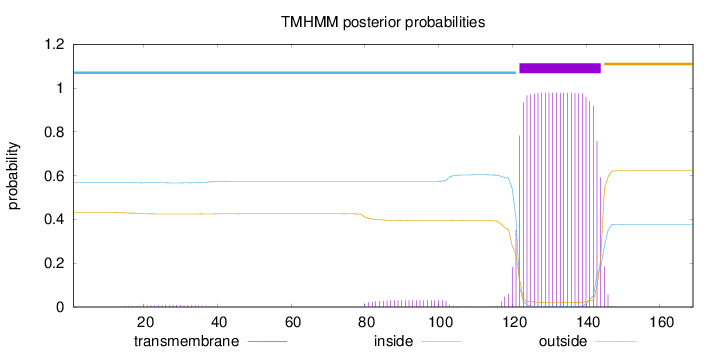
Length:
169
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.28057
Exp number, first 60 AAs:
0.16665
Total prob of N-in:
0.56753
inside
1 - 121
TMhelix
122 - 144
outside
145 - 169
Population Genetic Test Statistics
Pi
244.061991
Theta
191.361356
Tajima's D
0.817437
CLR
0.124303
CSRT
0.612569371531423
Interpretation
Uncertain
