Gene
KWMTBOMO15132
Pre Gene Modal
BGIBMGA004915
Annotation
PREDICTED:_gastrula_zinc_finger_protein_XlCGF57.1_isoform_X1_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 3.824
Sequence
CDS
ATGTGTAAAAAGTCCTGTGATGAGGACAAGTGGGAGAGTCACGTAGAAATCTGTCGTATCAGTTCTAATATTAAGCAACAATATTGCTGTAAAGAATGTGGCAAGAAGTTTAAGTTCAAAAAGAGCTATCTTCAGCACATAGAATTGAATCACTCCACATTACCTAATAGTGTTGCTTGCACAAAAAGTAAAATATGGTGTCCAAATGAAACTGTTCTGAAGGAACACATAGAGACAATACACAAAAGAGCTTCGTATGTATGCCCAGAATGCAACAAAGAATTTGTAAGACGTTCTCATGTAACTCGTCATATGACACAGAGTGGCTGTAATGGTCATCAGTTAAATTTGTATCCTTGTGAAATTTGCAACTCAAAATTCACAAGGAAAGATAATTTGATCGTACATCTCAGAACTCAACACATACATAAAAGTGATAAATACTTTTTTTGCAAGTCATGTACTTTTGAAACTAAAAACTTTTCTAGACTTGTAAATCATTGGCTTTTAAAACATTCAGGTACACCAAATTATTATGAATGCTATCATTGCGGTAAATCTACTGGTTCCCGAACAGCAATGGCCAAGCATTTAGAAATACACGGAGAAAAAAGCTACACCTGTGAAGTGTGCGGCTATGCTACATACACGGTAGAAGTAATGCGGCGTCACATCTTAACTCACGTTACGGATAAACCTTACAAATGCGAGCATTGTAACAGATCGTACATAAAGAAATCACAGCTCCAGCGACATTTAGACAATCATGATATGCAGAAAATGTGCCTTAAATGTGGAGAAACTTTCTCTACAACATGCCAATTGTTAGACCACAAAAACGTACACAAAGGAAATAGCAAATTCGCTTGTCATTACCAAGATTGCCGTTTCTCTAAGAAACCATTTTTAAGTGAGGCTCAATTGCAGAAACACGTAAAAGTTCACCTCGAAGATAGACCATACGAGTGCGAAGTGTGCGGCAAGCGGTTCGCGGTGGAGCTGCACATGCGGCGGCACCTGTCCACGCACACACTGGAGCGGCCGCGGCGCTGCATGTACTGCGTGTCGGCGCGCGCCTACGTGCGAGGCGAGAAGCTCGTAAAGCACGTGCGCGTGCACCACCCCGCCGTGTTCGGCGCGCACCTCAAGCACGTGCGTGCGGTCCTCGGTATCGAAGCGAATACGGTGAGAGTGAAGAAGTCGGAACTAGAGTGTATATTGAACATGTTGGACGCGGAGTCGGATCGGATCATGGACGGCTACGGGGGGACCGGCGTGTTGTACGGCGGCATGCAGGAACAAAAAGATGTCAACGAAAACAGTGACGAAAACATCTCTACGCAAAACGATAATGGACCTTTGATGACAGAAGAGGAGCTTTTGGACAACTTAGGGAAAATGCTGTTCAAACTGATTGATTCAGACACACTCGAGTGTTTCGGTTGGCCCGACGAGCCAGTTGATGTGGTGTTAGAGAAAGTGATCGAGCACTGCGGAGCTAAACCAGCGGATCGCACGATGTGGACGCGCATGCAACGCCTCCGAGAGAACACGAAGCATCTCTTCCTGTACGTGATAGACGACAAGAACATCGCCCGCATGCTGGACACGCACACCATAGACCAGATCGTCAAACATATACTCGCCCAAGTCAGCGACACCGACAAATGA
Protein
MCKKSCDEDKWESHVEICRISSNIKQQYCCKECGKKFKFKKSYLQHIELNHSTLPNSVACTKSKIWCPNETVLKEHIETIHKRASYVCPECNKEFVRRSHVTRHMTQSGCNGHQLNLYPCEICNSKFTRKDNLIVHLRTQHIHKSDKYFFCKSCTFETKNFSRLVNHWLLKHSGTPNYYECYHCGKSTGSRTAMAKHLEIHGEKSYTCEVCGYATYTVEVMRRHILTHVTDKPYKCEHCNRSYIKKSQLQRHLDNHDMQKMCLKCGETFSTTCQLLDHKNVHKGNSKFACHYQDCRFSKKPFLSEAQLQKHVKVHLEDRPYECEVCGKRFAVELHMRRHLSTHTLERPRRCMYCVSARAYVRGEKLVKHVRVHHPAVFGAHLKHVRAVLGIEANTVRVKKSELECILNMLDAESDRIMDGYGGTGVLYGGMQEQKDVNENSDENISTQNDNGPLMTEEELLDNLGKMLFKLIDSDTLECFGWPDEPVDVVLEKVIEHCGAKPADRTMWTRMQRLRENTKHLFLYVIDDKNIARMLDTHTIDQIVKHILAQVSDTDK
Summary
Uniprot
A0A3S2M3F7
A0A212ETW2
A0A1E1WSR9
A0A1Z2RRK4
A0A2A4J228
A0A194RL13
+ More
A0A194PFU9 A0A1Y1LAK7 A0A2R7WTH6 A0A2J7R1K9 A0A067R6F8 A0A2S2NPB1 D6WQI5 U4U5I4 A0A1B6E3W8 J9K9X6 T1JG23 A0A0M8ZST6 A0A023F2W5 A0A1L8DIW0 A0A023F2E3 A0A154PH90 A0A195EH84 A0A224X9C5 A0A069DWF7 A0A151WRK3 A0A195AWD9 E0VUG3 A0A195FDR5 A0A2A3EQ55 A0A087ZZQ7 A0A195BY37 A0A158NHW9 E2AV88 A0A0A9XCE0 A0A0A9XMQ0 A0A146LDP0 A0A0K8SR27 A0A0A9YAA1 E9JB51 T1I0X7 A0A147BRV8 A0A131XTJ0 A0A1J1HLK0 A0A336MUV5
A0A194PFU9 A0A1Y1LAK7 A0A2R7WTH6 A0A2J7R1K9 A0A067R6F8 A0A2S2NPB1 D6WQI5 U4U5I4 A0A1B6E3W8 J9K9X6 T1JG23 A0A0M8ZST6 A0A023F2W5 A0A1L8DIW0 A0A023F2E3 A0A154PH90 A0A195EH84 A0A224X9C5 A0A069DWF7 A0A151WRK3 A0A195AWD9 E0VUG3 A0A195FDR5 A0A2A3EQ55 A0A087ZZQ7 A0A195BY37 A0A158NHW9 E2AV88 A0A0A9XCE0 A0A0A9XMQ0 A0A146LDP0 A0A0K8SR27 A0A0A9YAA1 E9JB51 T1I0X7 A0A147BRV8 A0A131XTJ0 A0A1J1HLK0 A0A336MUV5
Pubmed
EMBL
RSAL01000053
RVE50072.1
AGBW02012531
OWR44891.1
GDQN01001040
JAT90014.1
+ More
KY938831 KY963168 ASA46472.1 ASA46488.1 NWSH01003479 PCG66217.1 KQ460045 KPJ18232.1 KQ459604 KPI92261.1 GEZM01062258 GEZM01062257 JAV69918.1 KK855503 PTY22859.1 NEVH01008206 PNF34720.1 KK852669 KDR18870.1 GGMR01006396 MBY19015.1 KQ971354 EFA06069.1 KB631982 ERL87603.1 GEDC01004678 JAS32620.1 ABLF02040311 JH432192 KQ435863 KOX70410.1 GBBI01003027 JAC15685.1 GFDF01007810 JAV06274.1 GBBI01003257 JAC15455.1 KQ434905 KZC11225.1 KQ978957 KYN27232.1 GFTR01007501 JAW08925.1 GBGD01000862 JAC88027.1 KQ982813 KYQ50305.1 KQ976725 KYM76563.1 DS235786 EEB17019.1 KQ981636 KYN38805.1 KZ288194 PBC33895.1 KQ978501 KYM93574.1 ADTU01016095 GL443059 EFN62642.1 GBHO01025207 JAG18397.1 GBHO01025209 JAG18395.1 GDHC01013482 JAQ05147.1 GBRD01010584 JAG55240.1 GBHO01015063 GBRD01010586 JAG28541.1 JAG55238.1 GL770938 EFZ09943.1 ACPB03015537 GEGO01001943 JAR93461.1 GEFM01006271 JAP69525.1 CVRI01000010 CRK88909.1 UFQT01002773 SSX34046.1
KY938831 KY963168 ASA46472.1 ASA46488.1 NWSH01003479 PCG66217.1 KQ460045 KPJ18232.1 KQ459604 KPI92261.1 GEZM01062258 GEZM01062257 JAV69918.1 KK855503 PTY22859.1 NEVH01008206 PNF34720.1 KK852669 KDR18870.1 GGMR01006396 MBY19015.1 KQ971354 EFA06069.1 KB631982 ERL87603.1 GEDC01004678 JAS32620.1 ABLF02040311 JH432192 KQ435863 KOX70410.1 GBBI01003027 JAC15685.1 GFDF01007810 JAV06274.1 GBBI01003257 JAC15455.1 KQ434905 KZC11225.1 KQ978957 KYN27232.1 GFTR01007501 JAW08925.1 GBGD01000862 JAC88027.1 KQ982813 KYQ50305.1 KQ976725 KYM76563.1 DS235786 EEB17019.1 KQ981636 KYN38805.1 KZ288194 PBC33895.1 KQ978501 KYM93574.1 ADTU01016095 GL443059 EFN62642.1 GBHO01025207 JAG18397.1 GBHO01025209 JAG18395.1 GDHC01013482 JAQ05147.1 GBRD01010584 JAG55240.1 GBHO01015063 GBRD01010586 JAG28541.1 JAG55238.1 GL770938 EFZ09943.1 ACPB03015537 GEGO01001943 JAR93461.1 GEFM01006271 JAP69525.1 CVRI01000010 CRK88909.1 UFQT01002773 SSX34046.1
Proteomes
Pfam
Interpro
IPR036236
Znf_C2H2_sf
+ More
IPR013087 Znf_C2H2_type
IPR041697 Znf-C2H2_11
IPR004087 KH_dom
IPR022034 FXMRP1_C_core
IPR016197 Chromo-like_dom_sf
IPR041560 Tudor_FRX1
IPR040148 FMR1
IPR040472 FMRP_KH0
IPR004088 KH_dom_type_1
IPR008395 Agenet-like_dom
IPR036612 KH_dom_type_1_sf
IPR019787 Znf_PHD-finger
IPR001965 Znf_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR011011 Znf_FYVE_PHD
IPR019786 Zinc_finger_PHD-type_CS
IPR013087 Znf_C2H2_type
IPR041697 Znf-C2H2_11
IPR004087 KH_dom
IPR022034 FXMRP1_C_core
IPR016197 Chromo-like_dom_sf
IPR041560 Tudor_FRX1
IPR040148 FMR1
IPR040472 FMRP_KH0
IPR004088 KH_dom_type_1
IPR008395 Agenet-like_dom
IPR036612 KH_dom_type_1_sf
IPR019787 Znf_PHD-finger
IPR001965 Znf_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR011011 Znf_FYVE_PHD
IPR019786 Zinc_finger_PHD-type_CS
Gene 3D
ProteinModelPortal
A0A3S2M3F7
A0A212ETW2
A0A1E1WSR9
A0A1Z2RRK4
A0A2A4J228
A0A194RL13
+ More
A0A194PFU9 A0A1Y1LAK7 A0A2R7WTH6 A0A2J7R1K9 A0A067R6F8 A0A2S2NPB1 D6WQI5 U4U5I4 A0A1B6E3W8 J9K9X6 T1JG23 A0A0M8ZST6 A0A023F2W5 A0A1L8DIW0 A0A023F2E3 A0A154PH90 A0A195EH84 A0A224X9C5 A0A069DWF7 A0A151WRK3 A0A195AWD9 E0VUG3 A0A195FDR5 A0A2A3EQ55 A0A087ZZQ7 A0A195BY37 A0A158NHW9 E2AV88 A0A0A9XCE0 A0A0A9XMQ0 A0A146LDP0 A0A0K8SR27 A0A0A9YAA1 E9JB51 T1I0X7 A0A147BRV8 A0A131XTJ0 A0A1J1HLK0 A0A336MUV5
A0A194PFU9 A0A1Y1LAK7 A0A2R7WTH6 A0A2J7R1K9 A0A067R6F8 A0A2S2NPB1 D6WQI5 U4U5I4 A0A1B6E3W8 J9K9X6 T1JG23 A0A0M8ZST6 A0A023F2W5 A0A1L8DIW0 A0A023F2E3 A0A154PH90 A0A195EH84 A0A224X9C5 A0A069DWF7 A0A151WRK3 A0A195AWD9 E0VUG3 A0A195FDR5 A0A2A3EQ55 A0A087ZZQ7 A0A195BY37 A0A158NHW9 E2AV88 A0A0A9XCE0 A0A0A9XMQ0 A0A146LDP0 A0A0K8SR27 A0A0A9YAA1 E9JB51 T1I0X7 A0A147BRV8 A0A131XTJ0 A0A1J1HLK0 A0A336MUV5
PDB
5WJQ
E-value=1.40158e-30,
Score=333
Ontologies
GO
PANTHER
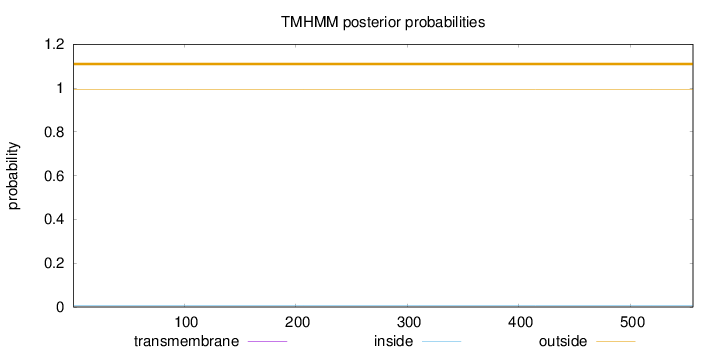
Topology
Length:
556
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00014
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00810
outside
1 - 556
Population Genetic Test Statistics
Pi
267.152984
Theta
210.708257
Tajima's D
1.464457
CLR
0.026002
CSRT
0.777661116944153
Interpretation
Uncertain
