Gene
KWMTBOMO14594
Pre Gene Modal
BGIBMGA008758
Annotation
PREDICTED:_zinc_finger_protein_454-like_[Amyelois_transitella]
Transcription factor
Location in the cell
Nuclear Reliability : 3.773
Sequence
CDS
ATGAGCATCCCGAAGGGCAAAGGTCCAATATTTGATCCGGGTTTGTGTCGATGTTGTGGTTCTATAAAGAAATGTCGTGTTCTGAATATAGAATACGAAAATAATGGGAAGAAAGAAGTTTATTCTGACATGATGATGGACTGCTTCGGCCTCTTACTGTCTCGCCTGGACGGGAAGCCATCTGAGAGGCTGGTCTGCGCCACATGCGTCATGAGAATCCGCGATGCCTTCTCCTTCCGACAGCAAGTACTCCACTGCGAAGAAGCCTTCTTACAAACGAAGGTGTATAATTCAACGAGTCTAGACTGTCAGTCTCTTCTAGAAGATGCGTTAGGGGACGGTGGCTCTCAAGTCGTTGTTGAAGTTGTTAAGACTGAACCCTGTGAGATGGATGAGCTTAGCATCGGCGGCGACATCTGTGCTGAGGTCTTACCTGACGTTAACGTCGAAGATACGAAGGCAGATCTAGGTTCGGATCACGATAGTTCAAACGACAACTTGCCATTAATACAACGCAGTGAGTGTAGAGAACCTAAGGAGGAAGTTCAGAGACCGATCACCAGGGCTAGTCAATTTAAACACTTGATGGGGAAGACGAATCGTATACCGATGACGAAGGTACAGCAGCTCATAAGAGAGAAAGACCCAACGTACATGACCGAAACGAACATATTGACCATAGTCGAATTCTCGTACGTGTGCCCGTTCAAATGTAGACACAATCACCTGCTCTGCTACTACTGCGGCGAGAACTTCTCGGATCCGCGTCTGATGCGGGAGCACACGATGAACCAGCACCACCCGAAGAAGTTTAAGGTGACCGAGCACAAGAACATGATCAAGGTGGATTTGACGAGGATCGACTGCAGACTGTGCCCTCAGAGGATCGACGATCTGGACGAGTTCAAGAGCCACATCGCTGGGACGCACAAGAAGAAGTACTACTTCGACTATAAGGACTCGGTGTTGCCGTTCCGTTTGACTAAGGAGGAGATCAGATGCGCGCTGTGCAACCTCGTGTTCCCTTACTTCCACGCTCTGAACAAACACATGAACGAGCATTTCAGTAATTACGTGTGCGAAACTTGCGGATTGGGTTTCGTGGACAAGGGCAGGTTCGTGATGCACCAGCAGCGCCACGAGCAGGGCGCCTTCCCCTGCGAGAGCTGCGGGAAGGTGTTCAAGGCGGAGTACAACCGGGACCTGCACGTGGACCGCGTGCACAGGAAGCGCGGCCGGGTCTACTGCCCGAAGTGCGACGTCAAGCTCATGTCCTACCCGCAGAAGCTGAAGCATCTGGTCGAGGTACACGGCGAGCAACCCATGTCGTTTCCTTGCACTATGTGCGATAAAGTATGCGAGACGCGACGCACCCTCACGATACACTGGAGGAAGGACCACTTGAAGGATTTCAGGTATAAAATGTTGGAAACGAACACAAAATGA
Protein
MSIPKGKGPIFDPGLCRCCGSIKKCRVLNIEYENNGKKEVYSDMMMDCFGLLLSRLDGKPSERLVCATCVMRIRDAFSFRQQVLHCEEAFLQTKVYNSTSLDCQSLLEDALGDGGSQVVVEVVKTEPCEMDELSIGGDICAEVLPDVNVEDTKADLGSDHDSSNDNLPLIQRSECREPKEEVQRPITRASQFKHLMGKTNRIPMTKVQQLIREKDPTYMTETNILTIVEFSYVCPFKCRHNHLLCYYCGENFSDPRLMREHTMNQHHPKKFKVTEHKNMIKVDLTRIDCRLCPQRIDDLDEFKSHIAGTHKKKYYFDYKDSVLPFRLTKEEIRCALCNLVFPYFHALNKHMNEHFSNYVCETCGLGFVDKGRFVMHQQRHEQGAFPCESCGKVFKAEYNRDLHVDRVHRKRGRVYCPKCDVKLMSYPQKLKHLVEVHGEQPMSFPCTMCDKVCETRRTLTIHWRKDHLKDFRYKMLETNTK
Summary
Uniprot
H9JGW1
A0A2A4JIU7
A0A212FGL8
A0A0N1IN28
A0A194R9B4
A0A2W1B758
+ More
H9JGV8 H9JGY5 A0A2H1W7P8 A0A2A4J4B8 A0A0N1IBB3 A0A194Q9V0 A0A212FGJ0 A0A194RAC3 A0A2H1VP71 A0A212FGI7 A0A2A4J945 S4NL96 A0A0N1PGX7 A0A0N0PDP7 H9JGV9 A0A2H1W1N2 A0A0N0P9X6 A0A3S2PBY9 H9JGY7 A0A3S2LYM8 A0A3S2NKW4 A0A2H1VCI6 A0A3S2NBW7 A0A2A4K5P2 A0A2W1BKF2 A0A2A4K7C4 H9JGU3 A0A194Q9X6 A0A2A4K655 A0A1E1WEJ1 A0A194R8P8 A0A2H1VCI8 A0A3S2NGJ4 A0A2A4K6K7 A0A0L7KVB5 H9JGX2 A0A0N1I342 A0A0N0PET9 H9JGX6 A0A2H1VPT0 A0A1E1W2N9 A0A1E1VY23 A0A1E1WUH1 A0A0N0P9L4 A0A2H1W1Q8 A0A3S2L6W4 H9JGZ7 A0A212FM36 A0A2H1W4Q3 A0A0N1PHE9 A0A2H1W2Z8 A0A0N0P9L1
H9JGV8 H9JGY5 A0A2H1W7P8 A0A2A4J4B8 A0A0N1IBB3 A0A194Q9V0 A0A212FGJ0 A0A194RAC3 A0A2H1VP71 A0A212FGI7 A0A2A4J945 S4NL96 A0A0N1PGX7 A0A0N0PDP7 H9JGV9 A0A2H1W1N2 A0A0N0P9X6 A0A3S2PBY9 H9JGY7 A0A3S2LYM8 A0A3S2NKW4 A0A2H1VCI6 A0A3S2NBW7 A0A2A4K5P2 A0A2W1BKF2 A0A2A4K7C4 H9JGU3 A0A194Q9X6 A0A2A4K655 A0A1E1WEJ1 A0A194R8P8 A0A2H1VCI8 A0A3S2NGJ4 A0A2A4K6K7 A0A0L7KVB5 H9JGX2 A0A0N1I342 A0A0N0PET9 H9JGX6 A0A2H1VPT0 A0A1E1W2N9 A0A1E1VY23 A0A1E1WUH1 A0A0N0P9L4 A0A2H1W1Q8 A0A3S2L6W4 H9JGZ7 A0A212FM36 A0A2H1W4Q3 A0A0N1PHE9 A0A2H1W2Z8 A0A0N0P9L1
EMBL
BABH01037786
BABH01037787
BABH01037788
NWSH01001274
PCG71891.1
AGBW02008655
+ More
OWR52853.1 KQ459348 KPJ01325.1 KQ460500 KPJ14222.1 KZ150324 PZC71398.1 BABH01037792 BABH01037793 BABH01037794 ODYU01006883 SOQ49125.1 NWSH01003163 PCG66841.1 KQ460130 KPJ17535.1 KQ459249 KPJ02298.1 OWR52854.1 KPJ14225.1 ODYU01003449 SOQ42242.1 OWR52852.1 NWSH01002560 PCG67980.1 GAIX01014736 JAA77824.1 KPJ01324.1 KPJ17537.1 BABH01037791 ODYU01005781 SOQ47000.1 KPJ01310.1 RSAL01000113 RVE47005.1 RVE47008.1 RSAL01000335 RVE42465.1 ODYU01001825 SOQ38559.1 RVE47007.1 NWSH01000101 PCG79547.1 KZ150113 PZC73316.1 PCG79550.1 BABH01037827 BABH01037828 BABH01037829 KPJ02297.1 PCG79549.1 GDQN01005632 JAT85422.1 KPJ14213.1 SOQ38560.1 RVE47011.1 PCG79548.1 JTDY01005417 KOB67026.1 BABH01037767 KPJ01321.1 KQ459805 KPJ19902.1 BABH01037765 BABH01037766 SOQ42244.1 GDQN01009814 JAT81240.1 GDQN01011465 JAT79589.1 GDQN01000613 JAT90441.1 KPJ01322.1 SOQ46998.1 RVE47010.1 BABH01037823 AGBW02007657 OWR54805.1 ODYU01006258 SOQ47936.1 KPJ17536.1 ODYU01006008 SOQ47480.1 KPJ01309.1
OWR52853.1 KQ459348 KPJ01325.1 KQ460500 KPJ14222.1 KZ150324 PZC71398.1 BABH01037792 BABH01037793 BABH01037794 ODYU01006883 SOQ49125.1 NWSH01003163 PCG66841.1 KQ460130 KPJ17535.1 KQ459249 KPJ02298.1 OWR52854.1 KPJ14225.1 ODYU01003449 SOQ42242.1 OWR52852.1 NWSH01002560 PCG67980.1 GAIX01014736 JAA77824.1 KPJ01324.1 KPJ17537.1 BABH01037791 ODYU01005781 SOQ47000.1 KPJ01310.1 RSAL01000113 RVE47005.1 RVE47008.1 RSAL01000335 RVE42465.1 ODYU01001825 SOQ38559.1 RVE47007.1 NWSH01000101 PCG79547.1 KZ150113 PZC73316.1 PCG79550.1 BABH01037827 BABH01037828 BABH01037829 KPJ02297.1 PCG79549.1 GDQN01005632 JAT85422.1 KPJ14213.1 SOQ38560.1 RVE47011.1 PCG79548.1 JTDY01005417 KOB67026.1 BABH01037767 KPJ01321.1 KQ459805 KPJ19902.1 BABH01037765 BABH01037766 SOQ42244.1 GDQN01009814 JAT81240.1 GDQN01011465 JAT79589.1 GDQN01000613 JAT90441.1 KPJ01322.1 SOQ46998.1 RVE47010.1 BABH01037823 AGBW02007657 OWR54805.1 ODYU01006258 SOQ47936.1 KPJ17536.1 ODYU01006008 SOQ47480.1 KPJ01309.1
Proteomes
Interpro
ProteinModelPortal
H9JGW1
A0A2A4JIU7
A0A212FGL8
A0A0N1IN28
A0A194R9B4
A0A2W1B758
+ More
H9JGV8 H9JGY5 A0A2H1W7P8 A0A2A4J4B8 A0A0N1IBB3 A0A194Q9V0 A0A212FGJ0 A0A194RAC3 A0A2H1VP71 A0A212FGI7 A0A2A4J945 S4NL96 A0A0N1PGX7 A0A0N0PDP7 H9JGV9 A0A2H1W1N2 A0A0N0P9X6 A0A3S2PBY9 H9JGY7 A0A3S2LYM8 A0A3S2NKW4 A0A2H1VCI6 A0A3S2NBW7 A0A2A4K5P2 A0A2W1BKF2 A0A2A4K7C4 H9JGU3 A0A194Q9X6 A0A2A4K655 A0A1E1WEJ1 A0A194R8P8 A0A2H1VCI8 A0A3S2NGJ4 A0A2A4K6K7 A0A0L7KVB5 H9JGX2 A0A0N1I342 A0A0N0PET9 H9JGX6 A0A2H1VPT0 A0A1E1W2N9 A0A1E1VY23 A0A1E1WUH1 A0A0N0P9L4 A0A2H1W1Q8 A0A3S2L6W4 H9JGZ7 A0A212FM36 A0A2H1W4Q3 A0A0N1PHE9 A0A2H1W2Z8 A0A0N0P9L1
H9JGV8 H9JGY5 A0A2H1W7P8 A0A2A4J4B8 A0A0N1IBB3 A0A194Q9V0 A0A212FGJ0 A0A194RAC3 A0A2H1VP71 A0A212FGI7 A0A2A4J945 S4NL96 A0A0N1PGX7 A0A0N0PDP7 H9JGV9 A0A2H1W1N2 A0A0N0P9X6 A0A3S2PBY9 H9JGY7 A0A3S2LYM8 A0A3S2NKW4 A0A2H1VCI6 A0A3S2NBW7 A0A2A4K5P2 A0A2W1BKF2 A0A2A4K7C4 H9JGU3 A0A194Q9X6 A0A2A4K655 A0A1E1WEJ1 A0A194R8P8 A0A2H1VCI8 A0A3S2NGJ4 A0A2A4K6K7 A0A0L7KVB5 H9JGX2 A0A0N1I342 A0A0N0PET9 H9JGX6 A0A2H1VPT0 A0A1E1W2N9 A0A1E1VY23 A0A1E1WUH1 A0A0N0P9L4 A0A2H1W1Q8 A0A3S2L6W4 H9JGZ7 A0A212FM36 A0A2H1W4Q3 A0A0N1PHE9 A0A2H1W2Z8 A0A0N0P9L1
PDB
2I13
E-value=9.61729e-05,
Score=110
Ontologies
GO
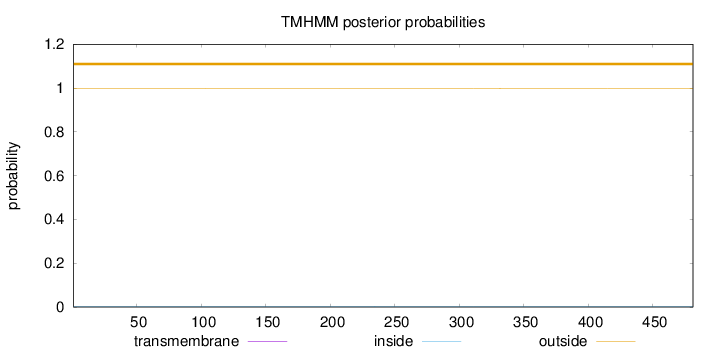
Topology
Length:
481
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0011
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00171
outside
1 - 481
Population Genetic Test Statistics
Pi
229.447837
Theta
170.772964
Tajima's D
2.060353
CLR
0.020223
CSRT
0.892755362231888
Interpretation
Uncertain
