Gene
KWMTBOMO14274
Annotation
PREDICTED:_zinc_finger_protein_431-like_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 2.97
Sequence
CDS
ATGTGGAAAAATTTCCTGAATAAGCTTGATTTCAGCTTGAATCTGTCTGAGACAACTAGAGCAAATGCACAAACAATATACATCATTATGGGAAAAAACAATACCATACTTAAAAGCAGAAAGAAGCAAGTGACAAACAAGAAAGAAGCATTGACTACAATTAAAGACAATATCAAAAGCAGACAGAAATATGTCAAGGTCAAAAACAAAAATGTCACCTGTGATGATTGCGGGGAGAGGTTCAAAGCAAAATGGATGTTAAAGCGACACATGAAGAACCGTTGTAGCACAAAATGTTCATGTCCTCAATGTCCCAAAGATTTTTCTACATATTCCTTATTAAAAGGACACATTGAAAGAATGCATCATCCAAAACGAATTAAATGCAAAAAATGCCCAAAAATGTTCAGCACTGAGAAATTACTACATAGACATGACAAAATGTACCACTTAGCAGCCATATGCAAACTGTGTTTTGTGCAATTCCCCTCTGGCAAAGATTTAAAAGTCCACTTAGACAAGCATGAAGTGTACAAATGTCCTCGTTGTGATAAATCACTATTGAATAAAAACACTTTGAAGTTTCATCTAAAAATATGTGGTAATGATGACTCAAAGAAATTAAGCTTTTTTTGTGACATATGTAAAAAAGGCTATGCACGTAAAAATGGTTTGCGAACACATTTGAAGACAGAACATGGGTTTGGTAACTCATTATCATGCAAATGGTGCAGTAAAAAGTTTGATGCTGTCAGTAGACTTAGGAATCACACTGTTAAACACACCAAAGAAAGAAATTATCACTGCGAGCATTGTGGGAACAGATTTGTGACACAAGCAGCTTTGGTTTATCATACCAGACTTCATACTGGTGAGAAACCATTTCCATGTGACATGTGCGAAGAAAGCTTTCTTTCAGCCTCTAGAAGAATGGAACATAAGAGAAGAAAACATTTAGGCCCTTCAAAAGAATGTCCCGTTTGCCACACAAAATTCATAACTACATACTGTTTGAAAAAGCACCTGGAAAGACATTATAATCCACAAAGTAAACTTTATATACTAGAGCCTGATATAAATATTGATCATCTTAGTGTGAATGATGTAAAACAATCTATGTATTTATTACAAACCTTTATATGA
Protein
MWKNFLNKLDFSLNLSETTRANAQTIYIIMGKNNTILKSRKKQVTNKKEALTTIKDNIKSRQKYVKVKNKNVTCDDCGERFKAKWMLKRHMKNRCSTKCSCPQCPKDFSTYSLLKGHIERMHHPKRIKCKKCPKMFSTEKLLHRHDKMYHLAAICKLCFVQFPSGKDLKVHLDKHEVYKCPRCDKSLLNKNTLKFHLKICGNDDSKKLSFFCDICKKGYARKNGLRTHLKTEHGFGNSLSCKWCSKKFDAVSRLRNHTVKHTKERNYHCEHCGNRFVTQAALVYHTRLHTGEKPFPCDMCEESFLSASRRMEHKRRKHLGPSKECPVCHTKFITTYCLKKHLERHYNPQSKLYILEPDINIDHLSVNDVKQSMYLLQTFI
Summary
Uniprot
A0A2H1WYP6
A0A2A4J049
A0A0N1IPL4
A0A194QCR4
A0A212ENM2
A0A2H1VT92
+ More
A0A194QMY8 A0A1E1W5A4 A0A194RJK0 S4PDZ1 A0A0N1PH02 A0A2W1B5B3 A0A2A4J1T4 A0A212ENN1 A0A194Q6R1 A0A212ERF6 A0A2H1WYD7 A0A0N0PA51 A0A0N1IBY4 A0A0L7LNQ4 A0A212EJS4 A0A2A4KBE3 A0A1E1WS62 H9JRM3 A0A2H1VIT1 A0A2H1WDU9 A0A0N1ICA8 A0A3B4B0H1 A0A146PC70 A0A3P9I1R6 A0A3B3TZT9 A0A3B3C108 A0A315VD53 M3ZX72 A0A3P9PHN3 A0A087X964 A0A3B3YJL6 A0A3Q2P1T9 A0A194PIN9 A0A146ZJ35 A0A146PSK8 A0A0N0PE00 A0A3Q2E8S7 A0A1A8HMD0 A0A146P3U9 A0A3Q2Z8V3 A0A147B650 H9IWQ3 A0A146XDC7 A0A1A7X9B4 A0A2H1VGT5 A0A3S2M357 A0A0L8G119 C3Y9P1 H9GYF6 A0A1A8R6B3 A0A146SBA6 A0A1A8L891 A0A2U9AXE0 A0A2I4BYA2 A0A3Q3ADR5 A0A1A8IVA6 A0A1A8E9S3 A0A1A7Z9K5 A0A3N0Y6S5 V5IFB2 A0A3P8SSE5 A0A0L8G874 A0A2M3Z9S7 A0A2A4K9V1 A0A3P8T641 C4A0V9 A0A2A4K9N4 A0A146PC25 A0A2M4BLI1 A0A3Q1B2D3 A0A3Q4MWC2 A0A3B5LRY9 A0A0L8I658 A0A182UES1 A0A131XPW7 A0A3B4T555 A0A146SFC8 A0A2M4AG03 A0A182U0D1 Q7QJB0 A0A3B4WEQ4 A0A0G2KK90 A0A1S3R335 A0A182IQC6 A0A084WN30 A0A3P9L4Z7 A0A2M4AFI9
A0A194QMY8 A0A1E1W5A4 A0A194RJK0 S4PDZ1 A0A0N1PH02 A0A2W1B5B3 A0A2A4J1T4 A0A212ENN1 A0A194Q6R1 A0A212ERF6 A0A2H1WYD7 A0A0N0PA51 A0A0N1IBY4 A0A0L7LNQ4 A0A212EJS4 A0A2A4KBE3 A0A1E1WS62 H9JRM3 A0A2H1VIT1 A0A2H1WDU9 A0A0N1ICA8 A0A3B4B0H1 A0A146PC70 A0A3P9I1R6 A0A3B3TZT9 A0A3B3C108 A0A315VD53 M3ZX72 A0A3P9PHN3 A0A087X964 A0A3B3YJL6 A0A3Q2P1T9 A0A194PIN9 A0A146ZJ35 A0A146PSK8 A0A0N0PE00 A0A3Q2E8S7 A0A1A8HMD0 A0A146P3U9 A0A3Q2Z8V3 A0A147B650 H9IWQ3 A0A146XDC7 A0A1A7X9B4 A0A2H1VGT5 A0A3S2M357 A0A0L8G119 C3Y9P1 H9GYF6 A0A1A8R6B3 A0A146SBA6 A0A1A8L891 A0A2U9AXE0 A0A2I4BYA2 A0A3Q3ADR5 A0A1A8IVA6 A0A1A8E9S3 A0A1A7Z9K5 A0A3N0Y6S5 V5IFB2 A0A3P8SSE5 A0A0L8G874 A0A2M3Z9S7 A0A2A4K9V1 A0A3P8T641 C4A0V9 A0A2A4K9N4 A0A146PC25 A0A2M4BLI1 A0A3Q1B2D3 A0A3Q4MWC2 A0A3B5LRY9 A0A0L8I658 A0A182UES1 A0A131XPW7 A0A3B4T555 A0A146SFC8 A0A2M4AG03 A0A182U0D1 Q7QJB0 A0A3B4WEQ4 A0A0G2KK90 A0A1S3R335 A0A182IQC6 A0A084WN30 A0A3P9L4Z7 A0A2M4AFI9
Pubmed
EMBL
ODYU01012014
SOQ58086.1
NWSH01004191
PCG65371.1
KQ460423
KPJ14825.1
+ More
KQ459386 KPJ01231.1 AGBW02013627 OWR43090.1 ODYU01004306 SOQ44029.1 KQ458859 KPJ04846.1 GDQN01008891 JAT82163.1 KQ460124 KPJ17614.1 GAIX01003431 JAA89129.1 KPJ14824.1 KZ150322 PZC71409.1 PCG65372.1 OWR43089.1 KPJ01232.1 AGBW02013061 OWR44034.1 SOQ58085.1 KQ458863 KPJ04728.1 KQ459988 KPJ18801.1 JTDY01000510 KOB76846.1 AGBW02014393 OWR41762.1 NWSH01000005 PCG80992.1 GDQN01001240 JAT89814.1 BABH01031739 ODYU01002539 SOQ40164.1 ODYU01007672 SOQ50664.1 KPJ04729.1 GCES01144910 JAQ41412.1 NHOQ01001935 PWA20668.1 AYCK01005444 KQ459602 KPI93187.1 GCES01019949 JAR66374.1 GCES01139595 JAQ46727.1 KPJ18802.1 HAEB01000267 HAEC01015578 SBQ83799.1 GCES01147830 JAQ38492.1 GCES01000045 JAR86278.1 BABH01021013 GCES01046262 JAR40061.1 HADW01012999 HADX01014946 SBP14399.1 ODYU01002490 SOQ40036.1 RSAL01000051 RVE50218.1 KQ424672 KOF70613.1 GG666492 EEN63297.1 CR450780 HAEH01014707 HAEI01015056 SBS00804.1 GCES01108672 JAQ77650.1 HAEF01003470 HAEG01000521 SBR40852.1 CP026243 AWO96324.1 HAED01014845 HAEE01005457 SBR01290.1 HADZ01006944 HAEA01013374 SBQ41854.1 HADY01001002 HAEJ01018466 SBP39487.1 RJVU01051426 ROL41889.1 GANP01008742 JAB75726.1 KQ423338 KOF73084.1 GGFM01004520 MBW25271.1 NWSH01000035 PCG80442.1 GG666831 EEN41572.1 PCG80440.1 GCES01144909 JAQ41413.1 GGFJ01004507 MBW53648.1 KQ416444 KOF96957.1 GEFM01006352 JAP69444.1 GCES01107064 JAQ79258.1 GGFK01006237 MBW39558.1 AAAB01008807 EAA04381.4 BX088603 ATLV01024557 KE525352 KFB51624.1 GGFK01006238 MBW39559.1
KQ459386 KPJ01231.1 AGBW02013627 OWR43090.1 ODYU01004306 SOQ44029.1 KQ458859 KPJ04846.1 GDQN01008891 JAT82163.1 KQ460124 KPJ17614.1 GAIX01003431 JAA89129.1 KPJ14824.1 KZ150322 PZC71409.1 PCG65372.1 OWR43089.1 KPJ01232.1 AGBW02013061 OWR44034.1 SOQ58085.1 KQ458863 KPJ04728.1 KQ459988 KPJ18801.1 JTDY01000510 KOB76846.1 AGBW02014393 OWR41762.1 NWSH01000005 PCG80992.1 GDQN01001240 JAT89814.1 BABH01031739 ODYU01002539 SOQ40164.1 ODYU01007672 SOQ50664.1 KPJ04729.1 GCES01144910 JAQ41412.1 NHOQ01001935 PWA20668.1 AYCK01005444 KQ459602 KPI93187.1 GCES01019949 JAR66374.1 GCES01139595 JAQ46727.1 KPJ18802.1 HAEB01000267 HAEC01015578 SBQ83799.1 GCES01147830 JAQ38492.1 GCES01000045 JAR86278.1 BABH01021013 GCES01046262 JAR40061.1 HADW01012999 HADX01014946 SBP14399.1 ODYU01002490 SOQ40036.1 RSAL01000051 RVE50218.1 KQ424672 KOF70613.1 GG666492 EEN63297.1 CR450780 HAEH01014707 HAEI01015056 SBS00804.1 GCES01108672 JAQ77650.1 HAEF01003470 HAEG01000521 SBR40852.1 CP026243 AWO96324.1 HAED01014845 HAEE01005457 SBR01290.1 HADZ01006944 HAEA01013374 SBQ41854.1 HADY01001002 HAEJ01018466 SBP39487.1 RJVU01051426 ROL41889.1 GANP01008742 JAB75726.1 KQ423338 KOF73084.1 GGFM01004520 MBW25271.1 NWSH01000035 PCG80442.1 GG666831 EEN41572.1 PCG80440.1 GCES01144909 JAQ41413.1 GGFJ01004507 MBW53648.1 KQ416444 KOF96957.1 GEFM01006352 JAP69444.1 GCES01107064 JAQ79258.1 GGFK01006237 MBW39558.1 AAAB01008807 EAA04381.4 BX088603 ATLV01024557 KE525352 KFB51624.1 GGFK01006238 MBW39559.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
UP000005204
+ More
UP000261520 UP000265200 UP000261500 UP000261560 UP000002852 UP000242638 UP000028760 UP000261480 UP000265000 UP000265020 UP000264820 UP000283053 UP000053454 UP000001554 UP000000437 UP000246464 UP000192220 UP000264800 UP000265080 UP000257160 UP000261580 UP000261380 UP000075902 UP000261420 UP000007062 UP000261360 UP000087266 UP000075880 UP000030765 UP000265180
UP000261520 UP000265200 UP000261500 UP000261560 UP000002852 UP000242638 UP000028760 UP000261480 UP000265000 UP000265020 UP000264820 UP000283053 UP000053454 UP000001554 UP000000437 UP000246464 UP000192220 UP000264800 UP000265080 UP000257160 UP000261580 UP000261380 UP000075902 UP000261420 UP000007062 UP000261360 UP000087266 UP000075880 UP000030765 UP000265180
Pfam
Interpro
ProteinModelPortal
A0A2H1WYP6
A0A2A4J049
A0A0N1IPL4
A0A194QCR4
A0A212ENM2
A0A2H1VT92
+ More
A0A194QMY8 A0A1E1W5A4 A0A194RJK0 S4PDZ1 A0A0N1PH02 A0A2W1B5B3 A0A2A4J1T4 A0A212ENN1 A0A194Q6R1 A0A212ERF6 A0A2H1WYD7 A0A0N0PA51 A0A0N1IBY4 A0A0L7LNQ4 A0A212EJS4 A0A2A4KBE3 A0A1E1WS62 H9JRM3 A0A2H1VIT1 A0A2H1WDU9 A0A0N1ICA8 A0A3B4B0H1 A0A146PC70 A0A3P9I1R6 A0A3B3TZT9 A0A3B3C108 A0A315VD53 M3ZX72 A0A3P9PHN3 A0A087X964 A0A3B3YJL6 A0A3Q2P1T9 A0A194PIN9 A0A146ZJ35 A0A146PSK8 A0A0N0PE00 A0A3Q2E8S7 A0A1A8HMD0 A0A146P3U9 A0A3Q2Z8V3 A0A147B650 H9IWQ3 A0A146XDC7 A0A1A7X9B4 A0A2H1VGT5 A0A3S2M357 A0A0L8G119 C3Y9P1 H9GYF6 A0A1A8R6B3 A0A146SBA6 A0A1A8L891 A0A2U9AXE0 A0A2I4BYA2 A0A3Q3ADR5 A0A1A8IVA6 A0A1A8E9S3 A0A1A7Z9K5 A0A3N0Y6S5 V5IFB2 A0A3P8SSE5 A0A0L8G874 A0A2M3Z9S7 A0A2A4K9V1 A0A3P8T641 C4A0V9 A0A2A4K9N4 A0A146PC25 A0A2M4BLI1 A0A3Q1B2D3 A0A3Q4MWC2 A0A3B5LRY9 A0A0L8I658 A0A182UES1 A0A131XPW7 A0A3B4T555 A0A146SFC8 A0A2M4AG03 A0A182U0D1 Q7QJB0 A0A3B4WEQ4 A0A0G2KK90 A0A1S3R335 A0A182IQC6 A0A084WN30 A0A3P9L4Z7 A0A2M4AFI9
A0A194QMY8 A0A1E1W5A4 A0A194RJK0 S4PDZ1 A0A0N1PH02 A0A2W1B5B3 A0A2A4J1T4 A0A212ENN1 A0A194Q6R1 A0A212ERF6 A0A2H1WYD7 A0A0N0PA51 A0A0N1IBY4 A0A0L7LNQ4 A0A212EJS4 A0A2A4KBE3 A0A1E1WS62 H9JRM3 A0A2H1VIT1 A0A2H1WDU9 A0A0N1ICA8 A0A3B4B0H1 A0A146PC70 A0A3P9I1R6 A0A3B3TZT9 A0A3B3C108 A0A315VD53 M3ZX72 A0A3P9PHN3 A0A087X964 A0A3B3YJL6 A0A3Q2P1T9 A0A194PIN9 A0A146ZJ35 A0A146PSK8 A0A0N0PE00 A0A3Q2E8S7 A0A1A8HMD0 A0A146P3U9 A0A3Q2Z8V3 A0A147B650 H9IWQ3 A0A146XDC7 A0A1A7X9B4 A0A2H1VGT5 A0A3S2M357 A0A0L8G119 C3Y9P1 H9GYF6 A0A1A8R6B3 A0A146SBA6 A0A1A8L891 A0A2U9AXE0 A0A2I4BYA2 A0A3Q3ADR5 A0A1A8IVA6 A0A1A8E9S3 A0A1A7Z9K5 A0A3N0Y6S5 V5IFB2 A0A3P8SSE5 A0A0L8G874 A0A2M3Z9S7 A0A2A4K9V1 A0A3P8T641 C4A0V9 A0A2A4K9N4 A0A146PC25 A0A2M4BLI1 A0A3Q1B2D3 A0A3Q4MWC2 A0A3B5LRY9 A0A0L8I658 A0A182UES1 A0A131XPW7 A0A3B4T555 A0A146SFC8 A0A2M4AG03 A0A182U0D1 Q7QJB0 A0A3B4WEQ4 A0A0G2KK90 A0A1S3R335 A0A182IQC6 A0A084WN30 A0A3P9L4Z7 A0A2M4AFI9
PDB
5V3M
E-value=8.19028e-20,
Score=239
Ontologies
GO
Topology
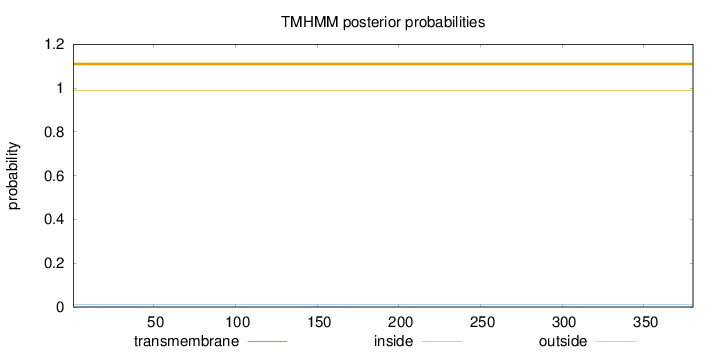
Length:
380
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00973
Exp number, first 60 AAs:
0.00617
Total prob of N-in:
0.01113
outside
1 - 380
Population Genetic Test Statistics
Pi
260.523221
Theta
192.87511
Tajima's D
1.185713
CLR
0.000091
CSRT
0.711914404279786
Interpretation
Uncertain
