Gene
KWMTBOMO14232
Pre Gene Modal
BGIBMGA011111
Annotation
PREDICTED:_zinc_finger_protein_OZF-like_isoform_X2_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 4.631
Sequence
CDS
ATGTCTAGTAGTTGGAACGGGTTTGGCCTTTGTCGTTGTTGTCATTCGAAAGGTAATTTCAAAACCCTCACTGCACCGCACACATATGAAGGGAAAACTGAAATGTATTCAGAGATATTGCTCAATACTTTTGAATTAAACGTGACTAAAAATTCCGACAGCCTAGATGCATATTGCATTTGTAGAAGATGCGAAACTAGACTCTTAGATGCTGCACAGTTCAAAAGACAAGTGCAGGAGTGTGAGCTTAATTTTTACCGCATTCTGGATTATAAAGATCTCCAAAATTTTGATGAATTGATAAAAAAAGAAAACGAAAGTACAGAATGTGAAGTGTTCAAATCGGAGTGTTTAGCAAATGAAAAAACTCACAATGACCACTCAGACTCCAAATTTGAAGACGATGAAAAACAATTTGATGCAGCCCAAGATCACTCTTCTGATTTTGAAGACACTCCCCTATCGGTGTTGCAGAGCATTAAAACGAACAGACAGACCGACCCCGCTGATTCGGAAGACAAAGAACCTGCTAAAATAGAAGATCTAAAACGAAACTGCGGAAAGGTGAAACCGAAAAAGTATTCGTGCGAGATATGCGAGAAGAGGTTCACGTACAAGGGGTCCCTGGAGGTGCACACGAGGCTGCACCGCGGCGAGGGCACGCTGGCCTGCCACCTGTGCGGCACCAAGCTGCCCGGCCACAAGGAGCTCACGCGCCACATCGCCGACACGCACGCCGTGGACGGCGGCTTCCCCTGCAACGCCTGCCCCGAGGTGTTCGACAAACCGCGCCTGCTCAAGCAGCACCTGCACGCGCACTACGAGCGGGACTTCTCCTGCGACGTCTGCTCCAAGGTGTTCCGCCGCAAGAAGGGCCTCAAGGAGCACCTGAAGACGCACACCGGCGAGAAGAAGTTCACGTGCGACATCTGCAGCAAGTCGTTCGGCCACAACCAGACGCTCAAGTCGCACGTGTACACGCACACGGGCGAGCGGCCGCACGTGTGCGACGTGTGCGGGCGCGGCTTCCCGCAGCGCACGCACCTGCGCCGCCACATCCGCGTGCTGCACCACCAGCTGCGCCCGCACGCCTGCGCCGTCTGCCACAAGCACTTCTCCAGCAAGAGCTACCTGCGCGTCCACGAGCGCCAGCACAGCGGCGCGCGCCCCTACTCCTGCGACCTCTGCCAGCGCAGCTTCGCCGCATACACGACCCTCAAGGTGCACATGCGCGTGCACACCGGCCTCAAGCCCTACTCCTGCGACTTCTGCGGCCGCCAGTTCGCGCAGATGACCAGCTTCAAGCTGCACCAGCGCACGCACACCGGCGAGAAGCCGTACACGTGTAATGTTTGCAAGAAACCGTTCGCCGATAACGGTTACTTGAAAATCCACTTGAGGATCCACACGGAGGAGAAGCCGTACTCCTGCGAGATCTGCAAGCGCTGTTTCCGCGAGACGGGCCAGCTCAAGCGGCACCTGCGCATCCACTCCGGGGTCAAGGCGTACACGTGCCGGATATGCAACAAGCAGATCGGGAACCTGTCCAAGCACATGCGCGTCCACTCCGACGTTAAGCCGTTTTCGTGCACGATATGCAATAAAGAGTTTTCCGTGAACGGCAACTTGAAGTTGCACATGCGGACGCACACAGGGCTCAGGCCATATAGCTGCGAGCTATGTCACAGTCACTTCACACAAAGCAGCGGACTCAAAAGACACCGCTGTGCGCACAAAAGTGACGTTTGA
Protein
MSSSWNGFGLCRCCHSKGNFKTLTAPHTYEGKTEMYSEILLNTFELNVTKNSDSLDAYCICRRCETRLLDAAQFKRQVQECELNFYRILDYKDLQNFDELIKKENESTECEVFKSECLANEKTHNDHSDSKFEDDEKQFDAAQDHSSDFEDTPLSVLQSIKTNRQTDPADSEDKEPAKIEDLKRNCGKVKPKKYSCEICEKRFTYKGSLEVHTRLHRGEGTLACHLCGTKLPGHKELTRHIADTHAVDGGFPCNACPEVFDKPRLLKQHLHAHYERDFSCDVCSKVFRRKKGLKEHLKTHTGEKKFTCDICSKSFGHNQTLKSHVYTHTGERPHVCDVCGRGFPQRTHLRRHIRVLHHQLRPHACAVCHKHFSSKSYLRVHERQHSGARPYSCDLCQRSFAAYTTLKVHMRVHTGLKPYSCDFCGRQFAQMTSFKLHQRTHTGEKPYTCNVCKKPFADNGYLKIHLRIHTEEKPYSCEICKRCFRETGQLKRHLRIHSGVKAYTCRICNKQIGNLSKHMRVHSDVKPFSCTICNKEFSVNGNLKLHMRTHTGLRPYSCELCHSHFTQSSGLKRHRCAHKSDV
Summary
Uniprot
EMBL
BABH01035209
ODYU01006399
SOQ48210.1
RSAL01000274
RVE43139.1
ODYU01003064
+ More
SOQ41364.1 AGBW02010319 OWR48738.1 AYCK01001272 AYCK01001273 ABLF02025528 KQ430326 KOF64322.1 AERX01021876 AERX01021877 AERX01021878 GG666612 EEN48846.1 ABLF02039175 ABLF02009695 ABLF02018750 ABLF02061073 ABLF02063523 GCES01007040 JAR79283.1
SOQ41364.1 AGBW02010319 OWR48738.1 AYCK01001272 AYCK01001273 ABLF02025528 KQ430326 KOF64322.1 AERX01021876 AERX01021877 AERX01021878 GG666612 EEN48846.1 ABLF02039175 ABLF02009695 ABLF02018750 ABLF02061073 ABLF02063523 GCES01007040 JAR79283.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF57667
SSF57667
ProteinModelPortal
PDB
5V3M
E-value=6.40359e-48,
Score=483
Ontologies
GO
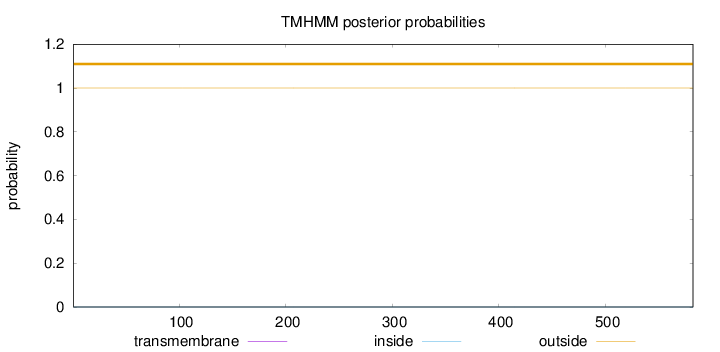
Topology
Length:
582
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00017
outside
1 - 582
Population Genetic Test Statistics
Pi
2.858074
Theta
6.189095
Tajima's D
-2.416763
CLR
1.037464
CSRT
0.000699965001749912
Interpretation
Possibly Positive selection
