Pre Gene Modal
BGIBMGA007172
Annotation
PREDICTED:_glycosylated_lysosomal_membrane_protein_A-like_[Bombyx_mori]
Transcription factor
Location in the cell
PlasmaMembrane Reliability : 2.491
Sequence
CDS
ATGAATTTCTTAATTTTATTGTTGAGTAGCGCGATATCTATAGTGTGTGGACAAGATCGAGTGATAACAGCGAAATTAAATCCGGGATGCGCCGAATGTACGTCAGCAAACACTTTAGTTTATATAAAGGCTGACAGCTCCAAGGACAGCATCCACCAGCTATGGGACTTCACAGGTGGGATCCCTACTGTGGTGTTTGCACTAACCGAACTGAACTCGACGATGCAAGTGAAATGGGACAGACAAGTGCCGGTGAAGTTTTTGTTGTCCGAAACACCAAAGTATTGCTTCGCTATTGCCATAGACAAGCTCTACGAATACAATGATGTGGAGGATAAGGGCCACATCAGCCCGCAGTGCGAACAGAGGCCGATGTCTCTCAAGTATATGTCGTGGACCCTGGTCGACAGCGTGCTCTCCGACAAGGAGGTGATGGTTCGAGTCCACGGTCAATATAAGCACAGCTGGCGAGCTGGCGTCGTGGATGTCAAGTTGGATCTCATCCCGTTCTACGACTACGCGGCCGAACTGCCCCGTCTCATACACACAGCAAACTCGACCCTCGTGGACGTTGGCCTCGTCAACATCACTACTTCGAAGGACTACAACTCGTCCCGTTTCGCCCTGCACATGCTGTTGGTCAGCACCGACGGATGGGGGGAGACCATGCACTACACCATGAGGAAGAGTCTCGACGACGAGCACACTCCAGGAGTTTTTGAGATCATCGAGATAAAGACGCCAGAGTCGTACGTGAAGGACGACGGCGGCTACATCCAGTTCCGGCCCGTGGGCTACACCGAACCGAAGCGCGAGGTCTCCTCGTCCACCTCCGTCCGAGTCTCCCCGCTCAAGAAGAGCGCCGTGCCGGAGCGCAGCACGCTGGAGCGTTTCTTCCGCGACTTCGAGACGGAATCTCTCCTGGTGCAAGACGTGTTCGTGTCGATCGGCCTCACCGGGGACCAGTTCTACAGACAACACAACTTTACCTCCTGGTCGTTCACGATGGGCTACGGCAGCCCGCCCCTAGAAACATTCTCGCTGTTCGTGATAATGATCATATCCATCGGCCTCGGGGTGCCCTTGTTGCTGGCGCTCTCTGGGGTCGTCTACGTGGTGATCTCCAGGATCAGACAGAGGAACTCCAACCAGCCGACCAGATTCACGGACGAGGAATGA
Protein
MNFLILLLSSAISIVCGQDRVITAKLNPGCAECTSANTLVYIKADSSKDSIHQLWDFTGGIPTVVFALTELNSTMQVKWDRQVPVKFLLSETPKYCFAIAIDKLYEYNDVEDKGHISPQCEQRPMSLKYMSWTLVDSVLSDKEVMVRVHGQYKHSWRAGVVDVKLDLIPFYDYAAELPRLIHTANSTLVDVGLVNITTSKDYNSSRFALHMLLVSTDGWGETMHYTMRKSLDDEHTPGVFEIIEIKTPESYVKDDGGYIQFRPVGYTEPKREVSSSTSVRVSPLKKSAVPERSTLERFFRDFETESLLVQDVFVSIGLTGDQFYRQHNFTSWSFTMGYGSPPLETFSLFVIMIISIGLGVPLLLALSGVVYVVISRIRQRNSNQPTRFTDEE
Summary
Similarity
Belongs to the class I-like SAM-binding methyltransferase superfamily. Trm1 family.
Uniprot
H9JCC7
A0A2H1V4V8
A0A1E1WLR1
A0A212FCR4
A0A2A4JVD7
A0A0N1INZ9
+ More
A0A437AWX3 A0A194QM05 A0A0L7LG30 B0XC61 A0A2J7QXI7 A0A195FAI2 A0A1Q3FEG9 A0A1Q3FEF8 A0A1B0DMW1 A0A2P8YLY4 Q0C746 E2BBX0 Q7QA54 A0A182NL39 A0A182MHS4 A0A154PP03 A0A182W3A1 E2AYQ8 A0A182XF84 A0A151IBW2 A0A1W7R7Z5 A0A182Y8Z0 A0A067QU12 A0A182QFW4 E9INK7 A0A023ERI9 A0A158NCE4 A0A195BWE2 A0A182PLI8 A0A1B6E8L9 A0A182T054 A0A1L8DTM7 F4WWY6 A0A182K9Y1 A0A1L8DTP7 A0A023ETR9 A0A151WMM4 A0A1L8DTI8 A0A1L8DTD8 A0A182KZQ9 A0A0J7KQV6 A0A0C9RLZ2 A0A182RI07 A0A0C9RFA9 A0A182VNM5 A0A0L7RG53 A0A182HVQ6 U5EN62 A0A182FQ65 A0A2M3Z6P8 A0A2M3Z6C2 A0A2M3ZDZ1 A0A2M3Z6D0 A0A2A3E7J3 A0A2M4BPP7 A0A2M4BSB0 A0A2M4BPW1 A0A2M4BQ09 A0A2M4ALB7 A0A087ZV47 W5JT17 A0A2J7QXG5 A0A1B6F7K0 A0A1B6H155 A0A1B6GQA5 A0A026VXB5 A0A1B6JGH5 A0A1B6IFE8 A0A224XHJ0 A0A182GHF1 K7IZT9 A0A232F7S5 E0VUP3 A0A1B0CH30 A0A1B0CDC9 T1I3Y6 A0A336LLA7 C3Y263 E9GU97 A0A1D2NJD3 A0A3R7QY22 A0A0P6DLS1 A0A0P5ZFF9 A0A0P5LWI9 A0A0M8ZV49 T1JFV0 A0A0P4XT94 A0A0N8ENG3
A0A437AWX3 A0A194QM05 A0A0L7LG30 B0XC61 A0A2J7QXI7 A0A195FAI2 A0A1Q3FEG9 A0A1Q3FEF8 A0A1B0DMW1 A0A2P8YLY4 Q0C746 E2BBX0 Q7QA54 A0A182NL39 A0A182MHS4 A0A154PP03 A0A182W3A1 E2AYQ8 A0A182XF84 A0A151IBW2 A0A1W7R7Z5 A0A182Y8Z0 A0A067QU12 A0A182QFW4 E9INK7 A0A023ERI9 A0A158NCE4 A0A195BWE2 A0A182PLI8 A0A1B6E8L9 A0A182T054 A0A1L8DTM7 F4WWY6 A0A182K9Y1 A0A1L8DTP7 A0A023ETR9 A0A151WMM4 A0A1L8DTI8 A0A1L8DTD8 A0A182KZQ9 A0A0J7KQV6 A0A0C9RLZ2 A0A182RI07 A0A0C9RFA9 A0A182VNM5 A0A0L7RG53 A0A182HVQ6 U5EN62 A0A182FQ65 A0A2M3Z6P8 A0A2M3Z6C2 A0A2M3ZDZ1 A0A2M3Z6D0 A0A2A3E7J3 A0A2M4BPP7 A0A2M4BSB0 A0A2M4BPW1 A0A2M4BQ09 A0A2M4ALB7 A0A087ZV47 W5JT17 A0A2J7QXG5 A0A1B6F7K0 A0A1B6H155 A0A1B6GQA5 A0A026VXB5 A0A1B6JGH5 A0A1B6IFE8 A0A224XHJ0 A0A182GHF1 K7IZT9 A0A232F7S5 E0VUP3 A0A1B0CH30 A0A1B0CDC9 T1I3Y6 A0A336LLA7 C3Y263 E9GU97 A0A1D2NJD3 A0A3R7QY22 A0A0P6DLS1 A0A0P5ZFF9 A0A0P5LWI9 A0A0M8ZV49 T1JFV0 A0A0P4XT94 A0A0N8ENG3
Pubmed
EMBL
BABH01031549
ODYU01000681
SOQ35871.1
GDQN01003105
JAT87949.1
AGBW02009157
+ More
OWR51498.1 NWSH01000508 PCG75971.1 KQ460882 KPJ11490.1 RSAL01000317 RVE42593.1 KQ458981 KPJ04486.1 JTDY01001247 KOB74412.1 DS232677 EDS44667.1 NEVH01009381 PNF33274.1 KQ981727 KYN37054.1 GFDL01009106 JAV25939.1 GFDL01009116 JAV25929.1 AJVK01017013 PYGN01000501 PSN45259.1 CH477186 EAT48981.1 GL447175 EFN86794.1 AAAB01008898 EAA09281.5 AXCM01001209 KQ435007 KZC13593.1 GL443983 EFN61421.1 KQ978076 KYM97211.1 GEHC01000410 JAV47235.1 KK853271 KDR09134.1 AXCN02000084 GL764306 EFZ17854.1 GAPW01001788 JAC11810.1 ADTU01011825 KQ976396 KYM92902.1 GEDC01003010 JAS34288.1 GFDF01004354 JAV09730.1 GL888417 EGI61229.1 GFDF01004293 JAV09791.1 GAPW01001784 JAC11814.1 KQ982934 KYQ49152.1 GFDF01004353 JAV09731.1 GFDF01004352 JAV09732.1 LBMM01004175 KMQ92733.1 GBYB01009255 JAG79022.1 GBYB01011782 JAG81549.1 KQ414598 KOC69813.1 APCN01005809 GANO01000721 JAB59150.1 GGFM01003430 MBW24181.1 GGFM01003318 MBW24069.1 GGFM01006023 MBW26774.1 GGFM01003257 MBW24008.1 KZ288348 PBC27474.1 GGFJ01005914 MBW55055.1 GGFJ01006537 MBW55678.1 GGFJ01005913 MBW55054.1 GGFJ01005912 MBW55053.1 GGFK01008264 MBW41585.1 ADMH02000531 ETN66080.1 PNF33275.1 GECZ01031637 GECZ01023573 GECZ01022185 JAS38132.1 JAS46196.1 JAS47584.1 GECZ01001357 JAS68412.1 GECZ01018562 GECZ01005168 GECZ01001707 JAS51207.1 JAS64601.1 JAS68062.1 KK107648 QOIP01000008 EZA48305.1 RLU19675.1 GECU01033059 GECU01028967 GECU01027725 GECU01020891 GECU01017616 GECU01014682 GECU01010067 GECU01009401 GECU01006986 JAS74647.1 JAS78739.1 JAS79981.1 JAS86815.1 JAS90090.1 JAS93024.1 JAS97639.1 JAS98305.1 JAT00721.1 GECU01022086 JAS85620.1 GFTR01005922 JAW10504.1 JXUM01063533 KQ562255 KXJ76300.1 NNAY01000704 OXU26891.1 DS235787 EEB17099.1 AJWK01011917 AJWK01007677 AJWK01007678 AJWK01007679 ACPB03003299 UFQT01000045 SSX18822.1 GG666480 EEN65953.1 GL732565 EFX77012.1 LJIJ01000030 ODN05076.1 QCYY01000789 ROT82686.1 GDIQ01139228 GDIQ01102756 GDIQ01090463 JAN04274.1 GDIP01047221 LRGB01002384 JAM56494.1 KZS07946.1 GDIQ01173177 JAK78548.1 KQ435830 KOX71755.1 JH432185 GDIP01238293 JAI85108.1 GDIQ01009564 JAN85173.1
OWR51498.1 NWSH01000508 PCG75971.1 KQ460882 KPJ11490.1 RSAL01000317 RVE42593.1 KQ458981 KPJ04486.1 JTDY01001247 KOB74412.1 DS232677 EDS44667.1 NEVH01009381 PNF33274.1 KQ981727 KYN37054.1 GFDL01009106 JAV25939.1 GFDL01009116 JAV25929.1 AJVK01017013 PYGN01000501 PSN45259.1 CH477186 EAT48981.1 GL447175 EFN86794.1 AAAB01008898 EAA09281.5 AXCM01001209 KQ435007 KZC13593.1 GL443983 EFN61421.1 KQ978076 KYM97211.1 GEHC01000410 JAV47235.1 KK853271 KDR09134.1 AXCN02000084 GL764306 EFZ17854.1 GAPW01001788 JAC11810.1 ADTU01011825 KQ976396 KYM92902.1 GEDC01003010 JAS34288.1 GFDF01004354 JAV09730.1 GL888417 EGI61229.1 GFDF01004293 JAV09791.1 GAPW01001784 JAC11814.1 KQ982934 KYQ49152.1 GFDF01004353 JAV09731.1 GFDF01004352 JAV09732.1 LBMM01004175 KMQ92733.1 GBYB01009255 JAG79022.1 GBYB01011782 JAG81549.1 KQ414598 KOC69813.1 APCN01005809 GANO01000721 JAB59150.1 GGFM01003430 MBW24181.1 GGFM01003318 MBW24069.1 GGFM01006023 MBW26774.1 GGFM01003257 MBW24008.1 KZ288348 PBC27474.1 GGFJ01005914 MBW55055.1 GGFJ01006537 MBW55678.1 GGFJ01005913 MBW55054.1 GGFJ01005912 MBW55053.1 GGFK01008264 MBW41585.1 ADMH02000531 ETN66080.1 PNF33275.1 GECZ01031637 GECZ01023573 GECZ01022185 JAS38132.1 JAS46196.1 JAS47584.1 GECZ01001357 JAS68412.1 GECZ01018562 GECZ01005168 GECZ01001707 JAS51207.1 JAS64601.1 JAS68062.1 KK107648 QOIP01000008 EZA48305.1 RLU19675.1 GECU01033059 GECU01028967 GECU01027725 GECU01020891 GECU01017616 GECU01014682 GECU01010067 GECU01009401 GECU01006986 JAS74647.1 JAS78739.1 JAS79981.1 JAS86815.1 JAS90090.1 JAS93024.1 JAS97639.1 JAS98305.1 JAT00721.1 GECU01022086 JAS85620.1 GFTR01005922 JAW10504.1 JXUM01063533 KQ562255 KXJ76300.1 NNAY01000704 OXU26891.1 DS235787 EEB17099.1 AJWK01011917 AJWK01007677 AJWK01007678 AJWK01007679 ACPB03003299 UFQT01000045 SSX18822.1 GG666480 EEN65953.1 GL732565 EFX77012.1 LJIJ01000030 ODN05076.1 QCYY01000789 ROT82686.1 GDIQ01139228 GDIQ01102756 GDIQ01090463 JAN04274.1 GDIP01047221 LRGB01002384 JAM56494.1 KZS07946.1 GDIQ01173177 JAK78548.1 KQ435830 KOX71755.1 JH432185 GDIP01238293 JAI85108.1 GDIQ01009564 JAN85173.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053240
UP000283053
UP000053268
+ More
UP000037510 UP000002320 UP000235965 UP000078541 UP000092462 UP000245037 UP000008820 UP000008237 UP000007062 UP000075884 UP000075883 UP000076502 UP000075920 UP000000311 UP000076407 UP000078542 UP000076408 UP000027135 UP000075886 UP000005205 UP000078540 UP000075885 UP000075901 UP000007755 UP000075881 UP000075809 UP000075882 UP000036403 UP000075900 UP000075903 UP000053825 UP000075840 UP000069272 UP000242457 UP000005203 UP000000673 UP000053097 UP000279307 UP000069940 UP000249989 UP000002358 UP000215335 UP000009046 UP000092461 UP000015103 UP000001554 UP000000305 UP000094527 UP000283509 UP000076858 UP000053105
UP000037510 UP000002320 UP000235965 UP000078541 UP000092462 UP000245037 UP000008820 UP000008237 UP000007062 UP000075884 UP000075883 UP000076502 UP000075920 UP000000311 UP000076407 UP000078542 UP000076408 UP000027135 UP000075886 UP000005205 UP000078540 UP000075885 UP000075901 UP000007755 UP000075881 UP000075809 UP000075882 UP000036403 UP000075900 UP000075903 UP000053825 UP000075840 UP000069272 UP000242457 UP000005203 UP000000673 UP000053097 UP000279307 UP000069940 UP000249989 UP000002358 UP000215335 UP000009046 UP000092461 UP000015103 UP000001554 UP000000305 UP000094527 UP000283509 UP000076858 UP000053105
PRIDE
Pfam
Interpro
Gene 3D
ProteinModelPortal
H9JCC7
A0A2H1V4V8
A0A1E1WLR1
A0A212FCR4
A0A2A4JVD7
A0A0N1INZ9
+ More
A0A437AWX3 A0A194QM05 A0A0L7LG30 B0XC61 A0A2J7QXI7 A0A195FAI2 A0A1Q3FEG9 A0A1Q3FEF8 A0A1B0DMW1 A0A2P8YLY4 Q0C746 E2BBX0 Q7QA54 A0A182NL39 A0A182MHS4 A0A154PP03 A0A182W3A1 E2AYQ8 A0A182XF84 A0A151IBW2 A0A1W7R7Z5 A0A182Y8Z0 A0A067QU12 A0A182QFW4 E9INK7 A0A023ERI9 A0A158NCE4 A0A195BWE2 A0A182PLI8 A0A1B6E8L9 A0A182T054 A0A1L8DTM7 F4WWY6 A0A182K9Y1 A0A1L8DTP7 A0A023ETR9 A0A151WMM4 A0A1L8DTI8 A0A1L8DTD8 A0A182KZQ9 A0A0J7KQV6 A0A0C9RLZ2 A0A182RI07 A0A0C9RFA9 A0A182VNM5 A0A0L7RG53 A0A182HVQ6 U5EN62 A0A182FQ65 A0A2M3Z6P8 A0A2M3Z6C2 A0A2M3ZDZ1 A0A2M3Z6D0 A0A2A3E7J3 A0A2M4BPP7 A0A2M4BSB0 A0A2M4BPW1 A0A2M4BQ09 A0A2M4ALB7 A0A087ZV47 W5JT17 A0A2J7QXG5 A0A1B6F7K0 A0A1B6H155 A0A1B6GQA5 A0A026VXB5 A0A1B6JGH5 A0A1B6IFE8 A0A224XHJ0 A0A182GHF1 K7IZT9 A0A232F7S5 E0VUP3 A0A1B0CH30 A0A1B0CDC9 T1I3Y6 A0A336LLA7 C3Y263 E9GU97 A0A1D2NJD3 A0A3R7QY22 A0A0P6DLS1 A0A0P5ZFF9 A0A0P5LWI9 A0A0M8ZV49 T1JFV0 A0A0P4XT94 A0A0N8ENG3
A0A437AWX3 A0A194QM05 A0A0L7LG30 B0XC61 A0A2J7QXI7 A0A195FAI2 A0A1Q3FEG9 A0A1Q3FEF8 A0A1B0DMW1 A0A2P8YLY4 Q0C746 E2BBX0 Q7QA54 A0A182NL39 A0A182MHS4 A0A154PP03 A0A182W3A1 E2AYQ8 A0A182XF84 A0A151IBW2 A0A1W7R7Z5 A0A182Y8Z0 A0A067QU12 A0A182QFW4 E9INK7 A0A023ERI9 A0A158NCE4 A0A195BWE2 A0A182PLI8 A0A1B6E8L9 A0A182T054 A0A1L8DTM7 F4WWY6 A0A182K9Y1 A0A1L8DTP7 A0A023ETR9 A0A151WMM4 A0A1L8DTI8 A0A1L8DTD8 A0A182KZQ9 A0A0J7KQV6 A0A0C9RLZ2 A0A182RI07 A0A0C9RFA9 A0A182VNM5 A0A0L7RG53 A0A182HVQ6 U5EN62 A0A182FQ65 A0A2M3Z6P8 A0A2M3Z6C2 A0A2M3ZDZ1 A0A2M3Z6D0 A0A2A3E7J3 A0A2M4BPP7 A0A2M4BSB0 A0A2M4BPW1 A0A2M4BQ09 A0A2M4ALB7 A0A087ZV47 W5JT17 A0A2J7QXG5 A0A1B6F7K0 A0A1B6H155 A0A1B6GQA5 A0A026VXB5 A0A1B6JGH5 A0A1B6IFE8 A0A224XHJ0 A0A182GHF1 K7IZT9 A0A232F7S5 E0VUP3 A0A1B0CH30 A0A1B0CDC9 T1I3Y6 A0A336LLA7 C3Y263 E9GU97 A0A1D2NJD3 A0A3R7QY22 A0A0P6DLS1 A0A0P5ZFF9 A0A0P5LWI9 A0A0M8ZV49 T1JFV0 A0A0P4XT94 A0A0N8ENG3
Ontologies
PANTHER
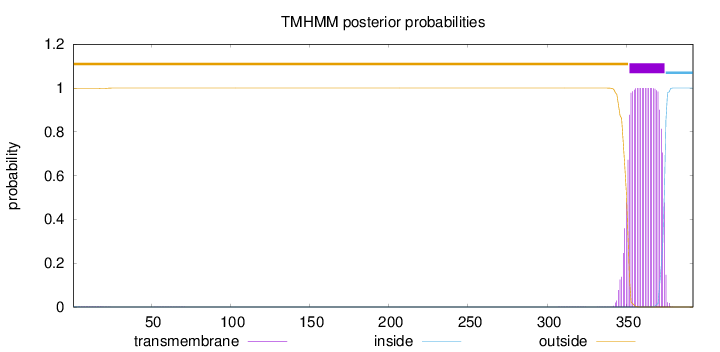
Topology
SignalP
Position: 1 - 17,
Likelihood: 0.994521
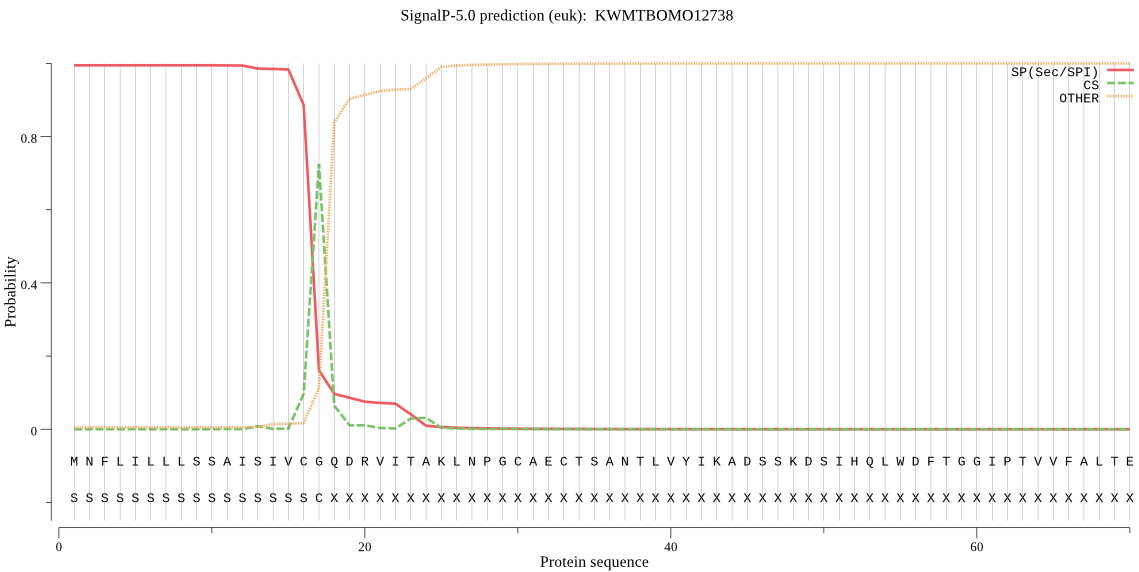
Length:
392
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
24.06996
Exp number, first 60 AAs:
0.02844
Total prob of N-in:
0.00157
outside
1 - 351
TMhelix
352 - 374
inside
375 - 392
Population Genetic Test Statistics
Pi
20.711349
Theta
20.050377
Tajima's D
0.14062
CLR
0.28325
CSRT
0.413179341032948
Interpretation
Uncertain
