Gene
KWMTBOMO12727
Pre Gene Modal
BGIBMGA007189
Annotation
PREDICTED:_zinc_finger_protein_845-like_[Plutella_xylostella]
Transcription factor
Location in the cell
Nuclear Reliability : 4.739
Sequence
CDS
ATGTACAGTCTACAGTCTAATCCATTACAAACTTATTTTGAAGAAACATTAGGAGTTAATCCTGTCTGTTCTATGGATTTGCCGCAGTGTGTGTGTTATGTATGTGCAGCATTATTGCAAAAATATAGTAGGTTCAAAACTAAGTGCTTACTTGGACAATCTGTGCTCTACCAAATACTAAACTCTGGTAAAAAGATTACAATCACATCAGTCCAAGAACTAGACCGAAGCAATTTAAAAACACCATTCGCCGTGTACACTGAAACAGATGTTAAAACAATTCACACAGTTTCCGACGATCCGAAGCTTCCAATTAAAGAAGAAATAGACATAGCGATAGATAGTAAAACATCATTAGAAAATGACCTGCAAGTGAATGAAATATTTCGTAAGGATGGCGAGTATGTGAAAGATGAGGATTACGGTGATTTCCCTTCTCAGCTAGAAGAGACAAGCGACGACGAACCGCTTTTAATGCACAAAATAAAGAAAGCTAAGAAAGCCAAAAAGAAAACGACCAAAAAGGATAAGAATAAAGTTGATTATATTTTGGCTGAAGACTTTCAAGGTGTCGAAGCAACAGACGATTCTTCGGCGTTACCACCAGAGATAACAGAAGTTTTACAGTCCAGAAAGAGAGGTCGACCAAGTAAAAAGAGAGTACAGGCAGAACCTGCGGGAAAGAAACGTAGAAGAACGAAAAATACGGGTGGGGTCACTGAAGACGATATTGTCTTGGAGGAATGTTGTACAATAACAACATTGAGTCAAGAAGAACAGCTGGAGGAGATCAACAAGCGGCAGTACTCCAGCAACTACATGAACGCTCAGTACAAGTGCCAGCTCTGTTACAAGGGCTTCATAGACACAGAGGCCTGGAGACATCACGTCTCCAAGCACTCACCCAGCGCGGGTGCAATAGAGTGTCCAGTGTGCAAGTTCCGCTTCGAGAACAAACGCCAGTTCCAGAAGCACGCCTCCAACCACGAGAAGAAGTACGCCTGCAACTACTGCGACCACGTCTCCGCCACGACAACCCAAGCGAAGCAGCACCAGCGCTGGCACAGGGGCGTCACGTACAAGTGCCAACACTGCGACGAGGTGTCCACCAAGTGGACGTCGTACCTGAGCCACGTACGCATGAAGCACCCCTCGGAGTTTATATGCGGCGCGTGCGGCTCCTCCTTCGTGAGCCGCCTGGGGCTGGCCATGCATAAGACCATGATGCACAAGGACCTGGATGAGAAGTCCGAAGCGGGCCCGTACTGCGCGGAGTGCGACGTCCAGTTCGCGTCGCTGGAGGCCTACAAGAGGCACATGGTCACGTCCGTCAAGCACGCCGAGCTCACGCTCACCAACAACGGCTGCCGGGTGTGCGGCGAGACGTTCGGCAGCGGGCACGAGCTGCGTGCGCACGCGCGGCGCCACCACGCCAAGAAGCGCCCCAAGAACTACGGGAAGAAGCCCACCAACCTCACTTGGCCCCTCAAGTGCGATCAGTGCTCACAAGAGGTGCATAGCGCGCGCGAGTACTGGAACCACTTCCGGCGGACCCATCCCGACAAGCCCTACCCCATTCAGAAGGACCACATCTGCGACGTGTGCGGAAAGAGCTTTAGGGGCAACGCGTTCCTGACGTACCACAAGCGCACCCACTCGGAGGAGCGGCAGTTCAAGTGCGAGACGTGCGGGAAGGCGTTCCACAACCCCGGCAACCTGTACATGCACGAGAAGATACACTCCGACCTCAGGCCCTACTCCTGCTCCGTCTGCTTCAAGGCCTTCAAGGGGAAGGGCGGACTCAGCAGACATTTTAGGTGTCACACCGGGGAGAGGCCTTACGATTGCGAGATCTGCGGCAAGGCGTTCAGTCAGTCGAACAGCCTCAAGGTGCACGTGCGCACGGTGCACTTAAAGCAGCCCGCGCCGTACGTCAGCCGCACCCGCCTGGAGAGGCGCCTCAAGGCCGGCACGGGCAAGGACAAACCTCACGAACCCTTCGCTTACACGTGA
Protein
MYSLQSNPLQTYFEETLGVNPVCSMDLPQCVCYVCAALLQKYSRFKTKCLLGQSVLYQILNSGKKITITSVQELDRSNLKTPFAVYTETDVKTIHTVSDDPKLPIKEEIDIAIDSKTSLENDLQVNEIFRKDGEYVKDEDYGDFPSQLEETSDDEPLLMHKIKKAKKAKKKTTKKDKNKVDYILAEDFQGVEATDDSSALPPEITEVLQSRKRGRPSKKRVQAEPAGKKRRRTKNTGGVTEDDIVLEECCTITTLSQEEQLEEINKRQYSSNYMNAQYKCQLCYKGFIDTEAWRHHVSKHSPSAGAIECPVCKFRFENKRQFQKHASNHEKKYACNYCDHVSATTTQAKQHQRWHRGVTYKCQHCDEVSTKWTSYLSHVRMKHPSEFICGACGSSFVSRLGLAMHKTMMHKDLDEKSEAGPYCAECDVQFASLEAYKRHMVTSVKHAELTLTNNGCRVCGETFGSGHELRAHARRHHAKKRPKNYGKKPTNLTWPLKCDQCSQEVHSAREYWNHFRRTHPDKPYPIQKDHICDVCGKSFRGNAFLTYHKRTHSEERQFKCETCGKAFHNPGNLYMHEKIHSDLRPYSCSVCFKAFKGKGGLSRHFRCHTGERPYDCEICGKAFSQSNSLKVHVRTVHLKQPAPYVSRTRLERRLKAGTGKDKPHEPFAYT
Summary
Uniprot
EMBL
RSAL01000080
RVE48654.1
KQ458981
KPJ04495.1
NWSH01000508
PCG75979.1
+ More
PCG75969.1 PCG75968.1 KQ460882 KPJ11499.1 RSAL01000098 RVE47649.1 RVE47650.1 NWSH01000652 PCG75007.1 NWSH01000946 PCG73425.1 KQ459463 KPJ00465.1 ODYU01007811 SOQ50892.1 KZ150164 PZC72671.1 PCG75008.1 ODYU01010673 SOQ55945.1 NWSH01000027 PCG80587.1 BABH01020992 BABH01020993 NWSH01000141 PCG79123.1 KZ149937 PZC77096.1 PZC77098.1 ABLF02027917 GCES01126360 JAQ59962.1
PCG75969.1 PCG75968.1 KQ460882 KPJ11499.1 RSAL01000098 RVE47649.1 RVE47650.1 NWSH01000652 PCG75007.1 NWSH01000946 PCG73425.1 KQ459463 KPJ00465.1 ODYU01007811 SOQ50892.1 KZ150164 PZC72671.1 PCG75008.1 ODYU01010673 SOQ55945.1 NWSH01000027 PCG80587.1 BABH01020992 BABH01020993 NWSH01000141 PCG79123.1 KZ149937 PZC77096.1 PZC77098.1 ABLF02027917 GCES01126360 JAQ59962.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF57667
SSF57667
Gene 3D
ProteinModelPortal
PDB
5V3M
E-value=1.39505e-25,
Score=291
Ontologies
GO
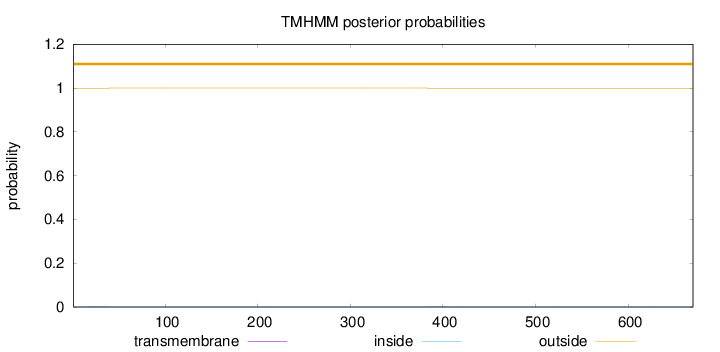
Topology
Length:
670
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01281
Exp number, first 60 AAs:
0.00629
Total prob of N-in:
0.00045
outside
1 - 670
Population Genetic Test Statistics
Pi
21.508726
Theta
20.077289
Tajima's D
0.072183
CLR
0.769671
CSRT
0.396830158492075
Interpretation
Uncertain
