Gene
KWMTBOMO12547
Pre Gene Modal
BGIBMGA001452
Annotation
reverse_transcriptase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.422 Nuclear Reliability : 2.1
Sequence
CDS
ATGAATAAGTCTCTGTTTGTACTCCTCGCGCTCGCCGCTGTCGCCTATGCCGACGAACCTCGCGGCTTAGATTTCAACCCTTGGACATGGCTATCGGCCACGAACAGAAACAAATCTCCCTCGATCATGGACTATTTCTTCCCACCGTCCCGAACGCCCAAACAATCAAACCTCGATCCGCCCGCCGATCGCAGGCCACCTCCTACCACGCAAAGTACGGACAGCACCCCGACAATGACGACCCTAGAAGAACGTACTACGTACGCGAAAAAAATCTCCACGAAAACTAAGTCGACTGTTCCGGTAACGAACCCTACAGCCACGACATCACCAAAAACCAAAAAGGGCACTCGGAAACCTACTACGACACATGAAACTAAAATAACGCAGAGTAAAACGGAAACGATCCCGCCGGCTACGAAAGACGCGACCGCTACTACGTCGAGGCCTACAAAACATTACACAATCCGAACCATAGTTAGAACGACAGAAACCCCTACGGTTCAAGACGCAGACACGACACTGAATACGGATAGTACAGAAACAGTTTCCGCTGATACGGCTGGTATTACGGTAGATGGGCCTAAAGTTACTGCTTCGACGGCGAACATAGCAGACGAGACGAGATCCACGGACGCCGGTACCGCGAGCACCGTCACCCTGGACTCGACCACGCAAACGGAGACTACTATAGAACCGGCCGACAGTCGCGAAGGATATCTACCGCTGCGCGACAAGCCCCAACAAACTCAAGTCAAGATCAATGAGCAAAACAAAAAAGAGCTCGCCTCGGAGTTTTTGGCCTTCAAGGCCAATCACACTGCGAGCCCTCTCGAGGACTCCGCCGCGCTCGCTGCTCCTGCGTCGCCTGTACCTGCGTGCAGAGCTTCTGCATTGAGCACCGCAGCCTCTGTCGTGCCTGCCGCTCCCGTGTCGCCTATACTGGCGAGAAAAGCTGCTGCGTCGTCCGCCGTGACCATCGTTTCAGCTGAGCGATCATCCGCGGCCTCCGTCGCGCCCTCTAAAACACCTACACTTGCTCGTAGGTCGCCTGCACCCGCCTCCTCGTGCTCCGACTCTGACTCGGACATGGAGGTCGACCTCGCCCCCGCCTCATCGACGGATGGATTCACCCTGGTACAGAAGGGTAAGAAGCGTGCCGCGGAGTCTCGAGCTCCCGCGGCCGCTAAAATTAGCAAAGCCGTGAACGCGTCGCGCCCCCGCCCTCAGACTCCCGTTGCGCCCCCAGCCCGTGCCACTCCGTCGCCGCGTCCGGTGGCACAAAATAAAACCCAGACCCCTCCCCCGGTTATCCTTCAGGAGAAGGCAGCTTGGGATCGAGTTTGCCTGGCCCTTAAGGCCAAAAATATAAATTTCACGAATGCCCGTAACCTCGCGAACGGCATTCAAATTAAGGTTCAAACACCCGACGACCATAGGGCCCTCTCTTCTTACCTCCGTAAGGAGCGTATAAGTTTCCATACGTATACGCTCCAGGAGGAGCGCGAACTCCGCGTTGTAATACGCGGAATCCCTAAAGAGTTAGATGTAGAGCTCGTAAAAGCCGACCTGTTAGAACAAGGCCTACCAGTGAATTCTGTGCACCGTATGCACACCGGTCGCGGTAGGGAGCCATATAATATGGTTCTAGTCGCTCTCCAGCCTACCCCGAGGGAGACCCTACTTAAGCCCGCGCGCCGTGACCCTAAAATCGCGAACTATAACATGGTCAGGAACGACAGGCTCTCTGCCCGTGGTGGTGGTACCGTCATTTACTATAGAAGAGCCCTGCATTGCGTCCCGCTCGATCCTCCCGCGCTCGCTAATATCGAAGCATCAGTGTGCCGAATCTCACTGACGGGACACGCGCCGATCGTTATCGCGTCCGTTTATCTTCCACCGGATAAGATCGTTCTAAGCAGTGATATCGAGGCGCTGCTCGGTATGGGGAGCTCTGTCATTCTGGCGGGCGACCTAAATTGTAAACACATCAGGTGGAACTCACACACCACAACCCCGAATGGCAGGCGGCTTGACGCGTTAGTCGATGATCTCGCCTTCGATATCGTCGCTCCGCTAACCCCGACTCACTACCCGCTAAATATCGCGCATCGCCCGGATATACTCGACATAGCGTTATTAAAAAACGTAACTCTGCGCTTACACTCGATCGAAGTAGTTTCAGAGTTAGATTCAGACCACCGTCCCGTCGTTATGAAGCTCGGTCGCGCTCCCGATTCCGTTCCCGTCACGAGGACTGTGGTGGATTGGCACACGCTGGGCATCAGCCTGGCTGAATCTGATCCACCATCGCTCCCGTTTAACCCGGACTCTACCCCGTCTCCTCAGGATACCGCTGAAGCCATAGACATCTTAACGTCACACATCACCTCGACATTAGATAGGTCATCGAAACAAGTTGTAGCGGAGGACTTCCTTCACCGCTTCAAATTGTCCGACGATATTAGGGAACTCCTTAGAGCTAAGAACGCCTCGATACGCGCCTACGACAGGTATCCTACCGCGGAAAATCGTATTCGAATGCGTGCCCTACAACGCGACGTAAAGTCTCGCATCGCCGAAGTCCGAGATGCCAGATGGTCTGATTTCTTAGAAGGACTCGCGCCCTCCCAAAGGTCTTACTACCGCTTAGCTCGTACTCTCAAATCGGATACGGTAGTAACTATGCCCCCCCTCGTAGGCCCCTCAGGCCGACTCGCGGCGTTCGATGATGACGAAAAAGCAGAGCTGCTGGCCGATACATTGCAAACCCAGTGCACGCCCAGCACTCAATCCGTGGACCCTGTTCATGTAGAATTAGTAGACAGTGAGGTAGAACGCAGAGCCTCCTTGCCACCCTCTGATGCGTTACCACCCGTCACCCCGATGGAAGTTAAAGACTTGATCAAAGACCTACGTCCTCGCAAGGCTCCCGGTTCCGACGGTATATCCAACCGCGTTATTAAACTTCTACCCGTCCAACTCATCGTGATGTTGGCATCTATTTTCAATGCCGCTATGGCGAACTGTATCTTTCCCGCGGTGTGGAAAGAAGCGGACGTTATCGGCATACATAAACCCGGTAAACCAAAAAATCATCCGACGAGCTACCGCCCGATTAGCCTCCTCATGTCTCTAGGCAAACTGTATGAGCGTCTGCTCTACAAACGCCTCAGAGACTTCGTCTCATCCAAGGGCATTCTTATCGATGAACAATTCGGATTCCGTACAAATCACTCATGCGTTCAACAGGTGCACCGCCTCACGGAGCACATTCTTGTGGGGCTTAATCGACCAAAACCGTTATACACGGGAGCTCTCTTCTTCGACGTCGCAAAAGCGTTCGACAAAGTCTGGCACAATGGTTTGATTTTCAAACTATTCAACATGGGCGTGCCGGATAGTCTCGTGCTCATCATACGGGACTTCTTGTCGAACCGCTCTTTTCGATATCGAGTCGAGGGAACCCGCTCCTCCCCACGACCTCTCACAGCTGGAGTCCCGCAAGGCTCTGTCCTCTCACCCCTCCTATTTAGCTTATTCGTCAACGATATTCCCCGGTCGCCGCCGACCCATTTAGCTTTATTCGCCGACGACACGACTGTTTACTATTCTAGTAGAAATAAGTCCCTAATCGCGAAGAAGCTTCAGAGCGCAGCCCTAGCCCTAGGACAGTGGTTCCGAAAATGGCGCATAGACATCAACCCAGCGAAAAGTACTGCGGTGCTATTTCAGAGGGGAAGCTCCACACGGATTTCCTCCCGGATTAGGAGGAGGAATCTCACACCCCCGATTACTCTCTTTAGACAACCCATACCCTGGGCCAGGAAGGTCAAATACCTGGGCGTTACCCTGGATGCATCGATGACATTCCGCCCGCATATAAAATCAGTCCGTGACCGTGCCGCGTTTATTCTCGGTAGACTCTACCCCATGATCTGTAAGCGGAGTAAAATGTCCCTTCGGAACAAGGTGACACTTTACAAAACTTGCATAAGGCCCGTCATGACTTACGCGAGTGTGGTGTTCGCTCACGCGGCCCGCACACACATAGACACCCTCCAATCCCTACAATCCCGCTTTTGCAGGTTAGCTGTCGGGGCTCCGTGGTTCGTGAGGAACGTTGACCTACACGATGACCTGGGCCTCGAATCAATTCGGAAATACATGAAGTCAGCGTCGGAACGATACTTCGATAAGGCTATGCGTCATGATAATCGCCTTATCGTTGCCGCCGCTGACTACTCCCCGAATCCTGATCATGCAGGAGCCAGTCACCGTCGACGCCCTAGACACGTCCTTACGGATCCATCAGATCCAATAACCTTTGCATTAGATGCCTTCAGGTCTAATACTAGGGGCAGGCTTAGGGACCCCGGTAACCGTACTCGTCGAACTCGACAAAGAGGTCGACGTGCAACCTAA
Protein
MNKSLFVLLALAAVAYADEPRGLDFNPWTWLSATNRNKSPSIMDYFFPPSRTPKQSNLDPPADRRPPPTTQSTDSTPTMTTLEERTTYAKKISTKTKSTVPVTNPTATTSPKTKKGTRKPTTTHETKITQSKTETIPPATKDATATTSRPTKHYTIRTIVRTTETPTVQDADTTLNTDSTETVSADTAGITVDGPKVTASTANIADETRSTDAGTASTVTLDSTTQTETTIEPADSREGYLPLRDKPQQTQVKINEQNKKELASEFLAFKANHTASPLEDSAALAAPASPVPACRASALSTAASVVPAAPVSPILARKAAASSAVTIVSAERSSAASVAPSKTPTLARRSPAPASSCSDSDSDMEVDLAPASSTDGFTLVQKGKKRAAESRAPAAAKISKAVNASRPRPQTPVAPPARATPSPRPVAQNKTQTPPPVILQEKAAWDRVCLALKAKNINFTNARNLANGIQIKVQTPDDHRALSSYLRKERISFHTYTLQEERELRVVIRGIPKELDVELVKADLLEQGLPVNSVHRMHTGRGREPYNMVLVALQPTPRETLLKPARRDPKIANYNMVRNDRLSARGGGTVIYYRRALHCVPLDPPALANIEASVCRISLTGHAPIVIASVYLPPDKIVLSSDIEALLGMGSSVILAGDLNCKHIRWNSHTTTPNGRRLDALVDDLAFDIVAPLTPTHYPLNIAHRPDILDIALLKNVTLRLHSIEVVSELDSDHRPVVMKLGRAPDSVPVTRTVVDWHTLGISLAESDPPSLPFNPDSTPSPQDTAEAIDILTSHITSTLDRSSKQVVAEDFLHRFKLSDDIRELLRAKNASIRAYDRYPTAENRIRMRALQRDVKSRIAEVRDARWSDFLEGLAPSQRSYYRLARTLKSDTVVTMPPLVGPSGRLAAFDDDEKAELLADTLQTQCTPSTQSVDPVHVELVDSEVERRASLPPSDALPPVTPMEVKDLIKDLRPRKAPGSDGISNRVIKLLPVQLIVMLASIFNAAMANCIFPAVWKEADVIGIHKPGKPKNHPTSYRPISLLMSLGKLYERLLYKRLRDFVSSKGILIDEQFGFRTNHSCVQQVHRLTEHILVGLNRPKPLYTGALFFDVAKAFDKVWHNGLIFKLFNMGVPDSLVLIIRDFLSNRSFRYRVEGTRSSPRPLTAGVPQGSVLSPLLFSLFVNDIPRSPPTHLALFADDTTVYYSSRNKSLIAKKLQSAALALGQWFRKWRIDINPAKSTAVLFQRGSSTRISSRIRRRNLTPPITLFRQPIPWARKVKYLGVTLDASMTFRPHIKSVRDRAAFILGRLYPMICKRSKMSLRNKVTLYKTCIRPVMTYASVVFAHAARTHIDTLQSLQSRFCRLAVGAPWFVRNVDLHDDLGLESIRKYMKSASERYFDKAMRHDNRLIVAAADYSPNPDHAGASHRRRPRHVLTDPSDPITFALDAFRSNTRGRLRDPGNRTRRTRQRGRRAT
Summary
Uniprot
EMBL
Proteomes
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
Ontologies
GO
Topology
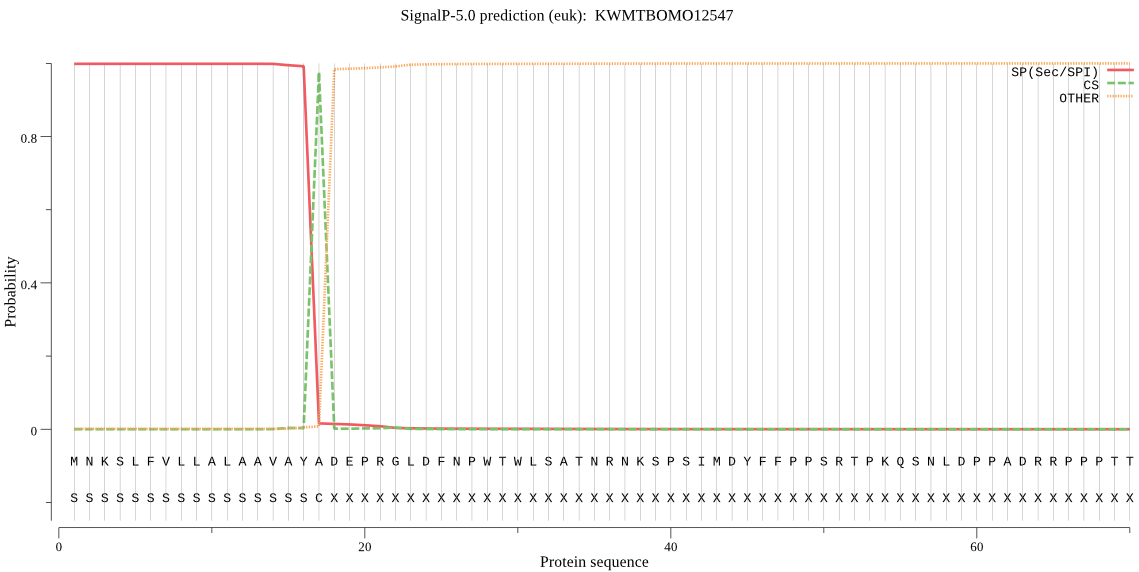
SignalP
Position: 1 - 17,
Likelihood: 0.998393
Length:
1474
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.15623
Exp number, first 60 AAs:
0.02421
Total prob of N-in:
0.00139
outside
1 - 1474

Population Genetic Test Statistics
Pi
148.083702
Theta
142.065964
Tajima's D
0.069719
CLR
103.333152
CSRT
0.390680465976701
Interpretation
Uncertain
