Gene
KWMTBOMO12252
Annotation
PREDICTED:_nascent_polypeptide-associated_complex_subunit_alpha?_muscle-specific_form-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.389
Sequence
CDS
ATGTCAGGGGGGAGCCCCCCCATAATCAGCACCCCAGGCCCCTCCCATAAGGGACGCACAATAGGAACTAGCCTATTTGCCCAAACCCAAACTCGCCCAGGCCCGGACAATAAGCCCGGGCCTTCTGGAGCGGCCCAGGGACCGCCGAGAATCGGTTCGGGGGAAAGTTGCTTTCCCTTACCGACCGATAACTCGGGGACCCCTCGGCCGCGAAAGAAGGCCAGGTCCGAGTCCGACCGGGGCTCTTCTGAGGAGAAAGACAAGAGACCGAGGATAGGTCCCTCTTCCAGATCCGAGGGTGACGATAGCGAAGCTACCTCCGTCACTCCTTCCTCTTCTGCATCCACATCTGCTTCGTCCTCCCCCTCCTCCTCTCGCTCAACGTCCCCCTCAAAGGAATCCCTTCCCTCACCCAAGAAACCCATCCCCAAACCTAGGGCACTCCCTAGGACGGTAGATGCTCCTAGGTTGAGAACCCCTCCGCGTTCTCCCTCTCCAGCACTCTCTCCTATCATGGCGGATAGTACTCGAGGTGCCCCCGCCTCAAGTTGCACGGCGGGCAACCTAGATTCAACTAGGTATGTGGACCTCGAGGCTACCCGACCCCCCCCTCCCTCAGTCCGAAAACTTCCTGACCCAATTCCGTCCACATCTTATGCGGCTATAGCCGCCTCCAAGCCAATTACTCCCAAGCCCCCAACTCAGGTCCATCGAAATGTTCCTCCCCCGAGACCGAAGGGTCGCGCCCAGGAGGAACTGCGCCGACACCCGCCAATAATGGTGGAGGCCCTTTCTAATTGGCCCCGCCACATGGCTGCCATTAGGGAGCGCCTAGGCCGCGCCCCCTCGGCGCGACCCTTCGGCACTGGTTTTAGGTTCCTGCCGTCGTCGGCTGAGGAATACAGGGCGGTCCAGGCCTACCTGTCGGAGGCCTCTGCCAGGGATAGTACCATTAAATGGTACTGCTATGCCCTGGCGGAGGAGATGCCCTCCAAGGTAGCTGTCCGGGGCCTCCCTGCCGACACCGACACGGCAGACGTTACCAACGCGTTGAAGGAGCTGGGGTTCCCGGCTCGATACGCTAGGTGCATTCGGTCCAAGAGAGGGCGACCGGGCAGCGTATTCCACGTTGCCCTGGATCACCTCTCAAAGGATGAACTCGCCCGCTTGTACTCAGTCAACGAACTGATGTACTTGCCGGGGGTGACGGTTGAGGGATGGCGACCAACCCGAGGGCCTGCGCAGTGTCACCGCTGCCAGGCCTACGGGCACGCCTCCCACAATTGTCACCGTGCGGTCCGCTGCGTGAGATGTGCTGCGGAGCACACTGTAGCGGACTGCCCGAGGCCGAGGGACGGCCCATTCTCCTGCGCCAATTGCGGGAAGGACCACGCCGCCGTTGATAGGCGGTGCCCGGTCTACCGCAAGAAGGCCAGGATGATGGGAGTAACCGTCCCTCCTCCGGTGCCACCGGTGCCACCGGTGCGTCGATCCGGGAATGCAGCTCCTTCCGCTGTAGCACCCACCTCGAAGGGGAAGGGGAAGGGGAAAGGGAAATCTTCCCAGCCCCAACCTCCTCCCACGTCCTCATTCATCCCCCCTGCCGTCACAGTGGAGGCCGAACCATCGGCGGCAACGCTGATGGCTGAAGCCAACACTCCGCGGCGGGGTAAACCTACTCCGACCCCACGTAGCTTCGCTATAACAAGACCGACCCCAAAACAAGACCGCGCTCCGGCCCGTGCCGGTGCCCCAACCCCACAAGCTCCAAATAAAAAAACCCTTAAAAACAGGAAAAAAAAGAGCAAAAGGAAATTGATGAAGAAAGCGCGAAGGGAGGCCTTAGAGAACGCTGCTCCCTCATCGGTTCCCGCAATACCTGCCATAGACATGGAGGTTGACATCAACCCCACTACCGACCACATAATCGATGATCAACAACCTGGACCATCTTCATTGCCCCAGGCCCAACCGGCCACTTCAGGCATACAGCCCCCATTACCACCCCGCCCGCAGAGGGAGCGGCGCAGCGGCCGACAGGACACGCAGCCTGCCGGCTTCGCTGCCTGGCTCCTCTCCCTGTGGGAGAAACTGTCCGAGGAGCTGCGCATCATCTACTGGAACCCCGACGGGATAGGGTCCCGGAGGTCGGAGCTCATCCGACTCGCCCAGGAGTATGACCCGCAGGTTATCCTCCTGGGCGAGACCAAGCTCAAACCTCGACAAGATTTTAGGATGCCCAACTACTTCGTGTACCGACGGGACGAACTGACCTCCAGCGGGCGCCCGTTCAGGGGCACTGCGGCGCTCGTCAGGAGGGACGTGGTGCACGAGGAGCTTGAGCTACTGACCTTCGCCTCTACCAGAACGGTCGGGGTGAGGGTCCACGTACCGGGGGCCGAATTGCGGATTTATGCCGCGTATCGACCCCCGGGTCCTGATTTTGTCGAGGACGATATTAGACGGGCCTTGAACCATGATCGCCATCCGGCCTTGGTGGTCGGGGACCTCAATGCGAAACATGTCGCTTGGGGCTCCCGAATCATCACGCCGCCCGGAAGGCGATTGTACGAGGATGCCGAAAGGGGGGACTACTGCGTGGTCGGCCCCAACGAACCCACACACGTTCCAACGTTGGCCCGGTACCAACCGGACGTATTGGACGTGTGTGTTCACAAGGGGCTACAGTGCCCTCTAGCGATAGAGGCACTGTACGACCTGAGTACCCCTCACCTACCGATCTTGGTCACGGTGGGTTTGGGGCTCACTCGGGTCGCGACATCCCCTCTCAGGCCTAGGGTCGACTGGCAGTCCTTTACGGGACTGCTGGCCGACCAATCCTTGTTTCAAGACCCGTCTACCCCTGATGAGGTCGACACGGCGGCTTCCCGTCTCGTCGACACCATCAGGGGAGCCCTGGACCAAGTGACGACTCTGGTACCCAGCGGGGGGCCCAGAGCGGAACTCCCGGTGCACCTCAAGACGTTAATTCGTCGGAAGAGGGCACTGAGGAGGCTCTGGGCGACATCTAGGTGTCCCAGGATTAAGCGCGAGCTCAACCGACTTGAGGGGGAGCTGGCGACTAAGATGCGGACTTACCGGGGTGATTCCTGGGAGGGGGGACTGTTGGACGCGTCCGACGACCGTACGATGGCCCCCCTTCACCGCATCTGTCGCCGGCTCACCAACAAGCCTTCCCCCACTTGCCCCTTGGAGGACGAGGGGGGAAGACGGTGTTTTACACCGTTGGCTCGTGCCGAAGTCCTGGCCAACTCGCTGGAGCGGCAGTTCACTCCGAATGTCTCTGCACCCTCGATGGCTGCATTCCATCGCGAGGTGTCAGAGGGTGTAATCCACTTCCTCAAACCGGAATCGCCGGGAGTCGAGCTGGGAGATGACGACTTAGTCACTCCCAGCGAGGTGCGCCTCCTCATCAAGTTGATGAAGAGGCGCAAAGCCCCCGGCGAGGACGGTATTCCTCCCCTCGCCCTGCAAAACCTCCCTCCCTCAATCGTGTCAGCAATGACACGATTGTTTAATTCCATTCTCCGGGTAGGACTCTTCCCTGGAACTTGGAAACTGGGCAAGATCATCGTCTTGCCCAAACCCGGCAAGGACAGGAGAAAGCCCTCCAGTTACCGACCGATTACCTTGCTCTCGCACGTGGGCAAGTTGTTCGAGCGGCTGCTCCTGAGGAGAATATCGCCGTATATTCTCCTCAGGCCGGAGCAATTCGGCTTTAGAAGCGGCCACTCGACGACTCTGCAACTGACCCGTGTCCTGCATCACTTGGCGGACCGGCGAAACTCGGGCCAATACACGGTCGCGGTCCTCCTCGATATCGAGAAGGCCTTCGACCGTGTGTGGCACGAGGGGCTGGTCCACAAGCTGCGGGACGCGGGCCTACCGCACGCTCACGTGCGACTGCTCGGGTCGTATCTGGAGGGTAGAACCTTCTTTGTCGCGGTCGAGGGCGCACGGTCTTCCGTGCGTCCCATTACGGCCGGGGTTCCCCAGGGTAGTGTCCTATCACCTATCCTATACGCCCTCTACACCAATGACATCCCCACCCTGGAGGGTCACCTTCGGGCGTGGGAGGAGGACGTGAAGTTGGCCTTGTTCGCTGACGACTCGGCCTACTTCTCGTCCTCTCCGTTCCCCTCTGCAGCCATTATGAGGATGCAGAGGCTTTTGGATTTACTGCCCCAGTGGCTGGACCGTTGGAGAGTCGCGGTCAATGTGGGAAAGACCGCGGCCTTACTCTACGGTCCGGTCCGTATCAGAGTCGTCCCAAAGAAACTCAGCCTCCGAGGTGTGAATGTCGAGTTCAGGACCACGGTCCGGTATCTCGGATGCGACATCGACCGCTCCTTGAGCATGGTCGCGCACGTCAACCGAGTCATCGGTCAGACTCGCGCGGCCAGGTTCTTGTTGCGGCCGGTGTTTGCGTCCGGGCTTCCGACCAGGACCAAACTCGCCATTTACAAGACATACGTCCGTTCCCGCCTCACATACGCTGCTGCGGCCTGGTATCACCTGTTGTCGCCGTCCCAGCGATCTAGGTTGCAGGCCCAGCAGAGTCTTTCCCTCCGCATTATCGTCGGGGCCGGGCAGTACGTCAGAAATGACGTTATTGCCCGGGACCTAGGTGTGGAGTCGCTCGTGGAGTTTGTGACTAGGCTCGCCCGTAATATGTACGAGCGAGCCGAAAACGGGCCCCATGAGCATCTCCACAACATAGCCCCGCTGCACGCCAGACCACCGGATGGTACGGCGTTGCCGCGGGAGCTATTGGTACCCCCGACGATCGTTCGCAGATAA
Protein
MSGGSPPIISTPGPSHKGRTIGTSLFAQTQTRPGPDNKPGPSGAAQGPPRIGSGESCFPLPTDNSGTPRPRKKARSESDRGSSEEKDKRPRIGPSSRSEGDDSEATSVTPSSSASTSASSSPSSSRSTSPSKESLPSPKKPIPKPRALPRTVDAPRLRTPPRSPSPALSPIMADSTRGAPASSCTAGNLDSTRYVDLEATRPPPPSVRKLPDPIPSTSYAAIAASKPITPKPPTQVHRNVPPPRPKGRAQEELRRHPPIMVEALSNWPRHMAAIRERLGRAPSARPFGTGFRFLPSSAEEYRAVQAYLSEASARDSTIKWYCYALAEEMPSKVAVRGLPADTDTADVTNALKELGFPARYARCIRSKRGRPGSVFHVALDHLSKDELARLYSVNELMYLPGVTVEGWRPTRGPAQCHRCQAYGHASHNCHRAVRCVRCAAEHTVADCPRPRDGPFSCANCGKDHAAVDRRCPVYRKKARMMGVTVPPPVPPVPPVRRSGNAAPSAVAPTSKGKGKGKGKSSQPQPPPTSSFIPPAVTVEAEPSAATLMAEANTPRRGKPTPTPRSFAITRPTPKQDRAPARAGAPTPQAPNKKTLKNRKKKSKRKLMKKARREALENAAPSSVPAIPAIDMEVDINPTTDHIIDDQQPGPSSLPQAQPATSGIQPPLPPRPQRERRSGRQDTQPAGFAAWLLSLWEKLSEELRIIYWNPDGIGSRRSELIRLAQEYDPQVILLGETKLKPRQDFRMPNYFVYRRDELTSSGRPFRGTAALVRRDVVHEELELLTFASTRTVGVRVHVPGAELRIYAAYRPPGPDFVEDDIRRALNHDRHPALVVGDLNAKHVAWGSRIITPPGRRLYEDAERGDYCVVGPNEPTHVPTLARYQPDVLDVCVHKGLQCPLAIEALYDLSTPHLPILVTVGLGLTRVATSPLRPRVDWQSFTGLLADQSLFQDPSTPDEVDTAASRLVDTIRGALDQVTTLVPSGGPRAELPVHLKTLIRRKRALRRLWATSRCPRIKRELNRLEGELATKMRTYRGDSWEGGLLDASDDRTMAPLHRICRRLTNKPSPTCPLEDEGGRRCFTPLARAEVLANSLERQFTPNVSAPSMAAFHREVSEGVIHFLKPESPGVELGDDDLVTPSEVRLLIKLMKRRKAPGEDGIPPLALQNLPPSIVSAMTRLFNSILRVGLFPGTWKLGKIIVLPKPGKDRRKPSSYRPITLLSHVGKLFERLLLRRISPYILLRPEQFGFRSGHSTTLQLTRVLHHLADRRNSGQYTVAVLLDIEKAFDRVWHEGLVHKLRDAGLPHAHVRLLGSYLEGRTFFVAVEGARSSVRPITAGVPQGSVLSPILYALYTNDIPTLEGHLRAWEEDVKLALFADDSAYFSSSPFPSAAIMRMQRLLDLLPQWLDRWRVAVNVGKTAALLYGPVRIRVVPKKLSLRGVNVEFRTTVRYLGCDIDRSLSMVAHVNRVIGQTRAARFLLRPVFASGLPTRTKLAIYKTYVRSRLTYAAAAWYHLLSPSQRSRLQAQQSLSLRIIVGAGQYVRNDVIARDLGVESLVEFVTRLARNMYERAENGPHEHLHNIAPLHARPPDGTALPRELLVPPTIVRR
Summary
Uniprot
ProteinModelPortal
PDB
6AR3
E-value=1.85595e-06,
Score=129
Ontologies
GO
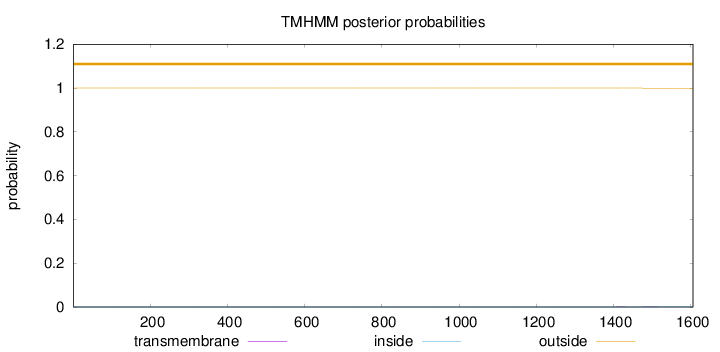
Topology
Length:
1606
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01607
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00000
outside
1 - 1606
Population Genetic Test Statistics
Pi
196.364226
Theta
170.503434
Tajima's D
0.260269
CLR
135.516925
CSRT
0.451627418629069
Interpretation
Uncertain
