Gene
KWMTBOMO11840
Annotation
PREDICTED:_zinc_finger_protein_808-like_isoform_X2_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 3.845
Sequence
CDS
ATGAGACAGCACTACACCAAATACGAGTGTCTGAGGTGTGACCTCGTGTGCGCGCAAGAGAGCACGATGCTGTATCACGAAGAATACCATAGTGGCGTGACAAGGAAATGCGTACGCTGCGACTTGGAGTTCAGACACGCGTCTACGTACTACTCGCACATGCGCACGCACCGCAGCGCGCACGTGTGCGTGTCGTGCGGCGCCTCGTACGTGAGCCGCGCCGGCCTGCAGCAGCACAGGAAGCTCAAGCACGTGCACGACGACGCGGACAGCCTCGCCGACGAGGACCGGGTCGCTTCGTACTGCGAAACGTGCGATGTCACGTTCGAATCCCGGCGGGCTTACGACGAACATCTATTTCACTCAGCCAAGCATTGTGACGACCAGTTGCAAGAGAAAAACGAAGCCATCGCTGTGTCAAGAAAGACTCTAAGCAAATCGTATAAGGCTAAAATGATCCCGATTCTAAAAAGACGGTGA
Protein
MRQHYTKYECLRCDLVCAQESTMLYHEEYHSGVTRKCVRCDLEFRHASTYYSHMRTHRSAHVCVSCGASYVSRAGLQQHRKLKHVHDDADSLADEDRVASYCETCDVTFESRRAYDEHLFHSAKHCDDQLQEKNEAIAVSRKTLSKSYKAKMIPILKRR
Summary
Uniprot
A0A2A4K4E2
A0A2A4K524
A0A0N1IQ78
A0A2H1WD86
A0A2W1BWP0
A0A2A4K5S6
+ More
A0A3S2NNE3 A0A212F6M7 A0A2H1V264 A0A3S2P4F7 A0A2W1BQ38 A0A0N1IHJ8 A0A2W1BV76 A0A0N1ING9 H9J4J0 A0A194QBF8 A0A2A4K8P9 A0A2W1BCB5 A0A2A4JTI5 A0A3S2LJV4 A0A194QQF7 H9IWW1 A0A194QAM2 A0A194Q527 A0A2W1BI44 A0A194QVL0 A0A212ET42 A0A3S2TJB5 H9JCD1 S4P6V7 A0A2A4JVY9 S4NXI2 A0A194Q632 S4PAC6 S4P6A2 A0A2H1VJG1 A0A212F5H6 A0A212EJ35 A0A2A4JSJ1 A0A2A4JVX9 A0A2A4JW96 A0A2H1W3X2 A0A1E1VZJ9 A0A2W1B1S3 A0A194QHT0 A0A2A4K955 A0A1E1WHH4 A0A0N1PJ21 A0A194QRI4 A0A2W1B689 A0A194QCN3 A0A2H1V5K9 A0A2H1WDF0 A0A2A4J6S1 A0A0L7LEH0 A0A194Q602 L7MDN9 A0A232F5R2 A0A0N1PIR8 K7JU69 A0A2A4JMU5 A0A0N1PHI2 A0A2Y9LJ83 S4PA08 A0A0N1IE84 W4XKC1 A0A194R1D7
A0A3S2NNE3 A0A212F6M7 A0A2H1V264 A0A3S2P4F7 A0A2W1BQ38 A0A0N1IHJ8 A0A2W1BV76 A0A0N1ING9 H9J4J0 A0A194QBF8 A0A2A4K8P9 A0A2W1BCB5 A0A2A4JTI5 A0A3S2LJV4 A0A194QQF7 H9IWW1 A0A194QAM2 A0A194Q527 A0A2W1BI44 A0A194QVL0 A0A212ET42 A0A3S2TJB5 H9JCD1 S4P6V7 A0A2A4JVY9 S4NXI2 A0A194Q632 S4PAC6 S4P6A2 A0A2H1VJG1 A0A212F5H6 A0A212EJ35 A0A2A4JSJ1 A0A2A4JVX9 A0A2A4JW96 A0A2H1W3X2 A0A1E1VZJ9 A0A2W1B1S3 A0A194QHT0 A0A2A4K955 A0A1E1WHH4 A0A0N1PJ21 A0A194QRI4 A0A2W1B689 A0A194QCN3 A0A2H1V5K9 A0A2H1WDF0 A0A2A4J6S1 A0A0L7LEH0 A0A194Q602 L7MDN9 A0A232F5R2 A0A0N1PIR8 K7JU69 A0A2A4JMU5 A0A0N1PHI2 A0A2Y9LJ83 S4PA08 A0A0N1IE84 W4XKC1 A0A194R1D7
EMBL
NWSH01000141
PCG79125.1
PCG79126.1
KQ459718
KPJ20255.1
ODYU01007868
+ More
SOQ51021.1 KZ149937 PZC77096.1 PCG79123.1 RSAL01000025 RVE52097.1 AGBW02009994 OWR49387.1 ODYU01000342 SOQ34930.1 RVE52099.1 PZC77098.1 LADI01012895 KPJ20750.1 PZC77097.1 KQ458408 KPJ05910.1 BABH01021778 KQ459460 KPJ00766.1 NWSH01000027 PCG80587.1 KZ150504 PZC70656.1 NWSH01000652 PCG75008.1 RSAL01000087 RVE48251.1 KQ461181 KPJ07589.1 BABH01021020 BABH01021021 BABH01021022 BABH01021023 KQ459463 KPJ00466.1 KPJ00464.1 KZ150333 PZC71343.1 KPJ07591.1 AGBW02012649 OWR44658.1 RSAL01000098 RVE47648.1 BABH01031546 GAIX01004849 JAA87711.1 NWSH01000508 PCG75979.1 GAIX01012157 JAA80403.1 KPJ00465.1 GAIX01006447 JAA86113.1 GAIX01005149 JAA87411.1 ODYU01002887 SOQ40936.1 AGBW02010195 OWR48987.1 AGBW02014523 OWR41513.1 PCG75007.1 PCG75969.1 PCG75968.1 ODYU01006174 SOQ47799.1 GDQN01010894 JAT80160.1 KZ150681 PZC70388.1 KQ458981 KPJ04495.1 PCG80589.1 GDQN01004610 JAT86444.1 KQ459941 KPJ19167.1 KPJ07590.1 KZ150686 PZC70385.1 KQ459193 KPJ03179.1 ODYU01000804 SOQ36135.1 ODYU01007811 SOQ50892.1 NWSH01002958 PCG67224.1 JTDY01001433 KOB73867.1 KQ459582 KPI98835.1 GACK01002688 JAA62346.1 NNAY01000861 OXU26166.1 KQ460140 KPJ17512.1 AAZX01002659 AAZX01018230 NWSH01000953 PCG73395.1 KQ459838 KPJ19752.1 GAIX01005246 JAA87314.1 KQ458725 KPJ05381.1 AAGJ04087397 KQ460878 KPJ11608.1
SOQ51021.1 KZ149937 PZC77096.1 PCG79123.1 RSAL01000025 RVE52097.1 AGBW02009994 OWR49387.1 ODYU01000342 SOQ34930.1 RVE52099.1 PZC77098.1 LADI01012895 KPJ20750.1 PZC77097.1 KQ458408 KPJ05910.1 BABH01021778 KQ459460 KPJ00766.1 NWSH01000027 PCG80587.1 KZ150504 PZC70656.1 NWSH01000652 PCG75008.1 RSAL01000087 RVE48251.1 KQ461181 KPJ07589.1 BABH01021020 BABH01021021 BABH01021022 BABH01021023 KQ459463 KPJ00466.1 KPJ00464.1 KZ150333 PZC71343.1 KPJ07591.1 AGBW02012649 OWR44658.1 RSAL01000098 RVE47648.1 BABH01031546 GAIX01004849 JAA87711.1 NWSH01000508 PCG75979.1 GAIX01012157 JAA80403.1 KPJ00465.1 GAIX01006447 JAA86113.1 GAIX01005149 JAA87411.1 ODYU01002887 SOQ40936.1 AGBW02010195 OWR48987.1 AGBW02014523 OWR41513.1 PCG75007.1 PCG75969.1 PCG75968.1 ODYU01006174 SOQ47799.1 GDQN01010894 JAT80160.1 KZ150681 PZC70388.1 KQ458981 KPJ04495.1 PCG80589.1 GDQN01004610 JAT86444.1 KQ459941 KPJ19167.1 KPJ07590.1 KZ150686 PZC70385.1 KQ459193 KPJ03179.1 ODYU01000804 SOQ36135.1 ODYU01007811 SOQ50892.1 NWSH01002958 PCG67224.1 JTDY01001433 KOB73867.1 KQ459582 KPI98835.1 GACK01002688 JAA62346.1 NNAY01000861 OXU26166.1 KQ460140 KPJ17512.1 AAZX01002659 AAZX01018230 NWSH01000953 PCG73395.1 KQ459838 KPJ19752.1 GAIX01005246 JAA87314.1 KQ458725 KPJ05381.1 AAGJ04087397 KQ460878 KPJ11608.1
Proteomes
Pfam
Interpro
IPR012934
Znf_AD
+ More
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR006612 THAP_Znf
IPR038441 THAP_Znf_sf
IPR022755 Znf_C2H2_jaz
IPR004686 Mtc
IPR001452 SH3_domain
IPR035755 RIM-BP_SH3_3
IPR013783 Ig-like_fold
IPR003961 FN3_dom
IPR040325 RIMBP1/2/3
IPR036116 FN3_sf
IPR035753 RIM-BP_SH3_2
IPR036028 SH3-like_dom_sf
IPR032714 DZIP1_N
IPR007246 Gaa1
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR006612 THAP_Znf
IPR038441 THAP_Znf_sf
IPR022755 Znf_C2H2_jaz
IPR004686 Mtc
IPR001452 SH3_domain
IPR035755 RIM-BP_SH3_3
IPR013783 Ig-like_fold
IPR003961 FN3_dom
IPR040325 RIMBP1/2/3
IPR036116 FN3_sf
IPR035753 RIM-BP_SH3_2
IPR036028 SH3-like_dom_sf
IPR032714 DZIP1_N
IPR007246 Gaa1
Gene 3D
ProteinModelPortal
A0A2A4K4E2
A0A2A4K524
A0A0N1IQ78
A0A2H1WD86
A0A2W1BWP0
A0A2A4K5S6
+ More
A0A3S2NNE3 A0A212F6M7 A0A2H1V264 A0A3S2P4F7 A0A2W1BQ38 A0A0N1IHJ8 A0A2W1BV76 A0A0N1ING9 H9J4J0 A0A194QBF8 A0A2A4K8P9 A0A2W1BCB5 A0A2A4JTI5 A0A3S2LJV4 A0A194QQF7 H9IWW1 A0A194QAM2 A0A194Q527 A0A2W1BI44 A0A194QVL0 A0A212ET42 A0A3S2TJB5 H9JCD1 S4P6V7 A0A2A4JVY9 S4NXI2 A0A194Q632 S4PAC6 S4P6A2 A0A2H1VJG1 A0A212F5H6 A0A212EJ35 A0A2A4JSJ1 A0A2A4JVX9 A0A2A4JW96 A0A2H1W3X2 A0A1E1VZJ9 A0A2W1B1S3 A0A194QHT0 A0A2A4K955 A0A1E1WHH4 A0A0N1PJ21 A0A194QRI4 A0A2W1B689 A0A194QCN3 A0A2H1V5K9 A0A2H1WDF0 A0A2A4J6S1 A0A0L7LEH0 A0A194Q602 L7MDN9 A0A232F5R2 A0A0N1PIR8 K7JU69 A0A2A4JMU5 A0A0N1PHI2 A0A2Y9LJ83 S4PA08 A0A0N1IE84 W4XKC1 A0A194R1D7
A0A3S2NNE3 A0A212F6M7 A0A2H1V264 A0A3S2P4F7 A0A2W1BQ38 A0A0N1IHJ8 A0A2W1BV76 A0A0N1ING9 H9J4J0 A0A194QBF8 A0A2A4K8P9 A0A2W1BCB5 A0A2A4JTI5 A0A3S2LJV4 A0A194QQF7 H9IWW1 A0A194QAM2 A0A194Q527 A0A2W1BI44 A0A194QVL0 A0A212ET42 A0A3S2TJB5 H9JCD1 S4P6V7 A0A2A4JVY9 S4NXI2 A0A194Q632 S4PAC6 S4P6A2 A0A2H1VJG1 A0A212F5H6 A0A212EJ35 A0A2A4JSJ1 A0A2A4JVX9 A0A2A4JW96 A0A2H1W3X2 A0A1E1VZJ9 A0A2W1B1S3 A0A194QHT0 A0A2A4K955 A0A1E1WHH4 A0A0N1PJ21 A0A194QRI4 A0A2W1B689 A0A194QCN3 A0A2H1V5K9 A0A2H1WDF0 A0A2A4J6S1 A0A0L7LEH0 A0A194Q602 L7MDN9 A0A232F5R2 A0A0N1PIR8 K7JU69 A0A2A4JMU5 A0A0N1PHI2 A0A2Y9LJ83 S4PA08 A0A0N1IE84 W4XKC1 A0A194R1D7
PDB
5V3G
E-value=0.0515655,
Score=80
Ontologies
GO
Topology
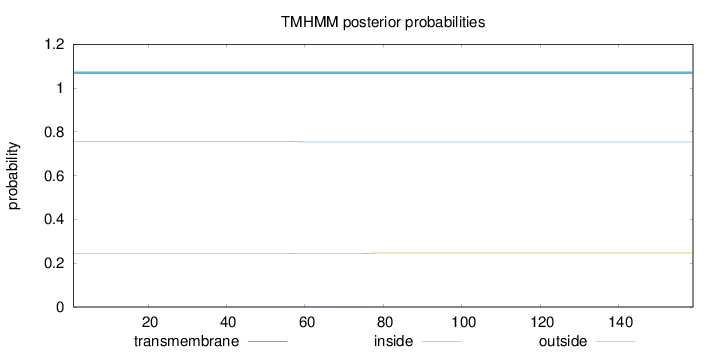
Length:
159
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03551
Exp number, first 60 AAs:
0.00484
Total prob of N-in:
0.75517
inside
1 - 159
Population Genetic Test Statistics
Pi
196.029373
Theta
164.117356
Tajima's D
0.851497
CLR
0.206471
CSRT
0.619869006549672
Interpretation
Uncertain
