Gene
KWMTBOMO11732
Pre Gene Modal
BGIBMGA004164
Annotation
PREDICTED:_transcription_factor_jun-D_[Bombyx_mori]
Transcription factor
Location in the cell
Mitochondrial Reliability : 2.067 Nuclear Reliability : 2.202
Sequence
CDS
ATGGTTCGGAATTCCGGCCACAGCATGGAGACCACCTTCTATGACGAGCAATATCCTCTCAGCGGCCCTGTGGAAAACCTTAAAAGGCCCCTAACCTTGGATGTTGGGCGGGGTGGGAAACGTTCACGAATCGCTGCACCCGTTCTCTCCTCGCCAGATTTACAAATGTTGAAGCTCGGATCTCCTGAGTTGGAAAAATTGATCATTCAAAACGGTATGGTGACAACCGCTACCCCTACTCCGGGAGCGCCTGTCTTATTTCCCGCAGTAGCACCTACGGAGGAACAAGAAATGTACGCGCGACCTTTCGTCGAAGCGCTAGACAAGCTGCACCACGGCGAGGCAGTGACGCCGCTCGGACGTCGAGTATATGCCGATTTGGACAGGCCGCTAGAGCGATACCCCACTCCCATCGTGAAGGATGAGCCCCAGACGGTGCCGAGCGCGGCCAGCACGCCGCCGCTATCACCCATCGACATGGACACTCAGGAAAAGATCAAACTGGAGAGAAAGAGACAGAGGAATCGGGTAGCCGCTTCCAAATGTAGACGACGTAAGCTGGAACGTATTTCAAAGCTGGAAGAGAAAGTGAAAATTCTTAAAGGTGAAAATGCTGAACTAGCACAGATGGTGGTGAAACTAAGGGACCACGTGCACAGGCTAAAGGAGCAGGTCCTAGAGCATGCGAATGGTGGCTGCCACATCGAGTCGCACTTCTGA
Protein
MVRNSGHSMETTFYDEQYPLSGPVENLKRPLTLDVGRGGKRSRIAAPVLSSPDLQMLKLGSPELEKLIIQNGMVTTATPTPGAPVLFPAVAPTEEQEMYARPFVEALDKLHHGEAVTPLGRRVYADLDRPLERYPTPIVKDEPQTVPSAASTPPLSPIDMDTQEKIKLERKRQRNRVAASKCRRRKLERISKLEEKVKILKGENAELAQMVVKLRDHVHRLKEQVLEHANGGCHIESHF
Summary
Similarity
Belongs to the bZIP family.
Uniprot
H9J3S7
A0A2H1V3B1
A0A2W1BIX9
I3UIH5
A0A2A4K553
A0A1E1WQD9
+ More
S4PA52 A0A0L7KW91 A0A194QY06 I4DMV0 I4DNB1 A0A3S2NJJ1 A0A067RC76 A0A2P8Y6Z1 A0A2J7QWY3 A0A3L8DJS6 A0A0J7L1Y1 A0A2R7VVD0 V5QNC4 A0A026WMR4 E2AK51 A0A195CU26 A0A195FSM8 A0A158NKX1 F4WAD7 A0A151I2C7 A0A151WMR4 A0A1W7HGV7 A0A195E3E4 A0A0P4VS28 R4G8D4 E9IYZ9 A0A224XWF8 A0A0V0G7D7 R4WCQ7 A0A069DRH1 A0A2R5LK36 A0A1B6DGV8 A0A310SYT0 A0A168GIS3 V9II37 A0A0M8ZYH5 A0A218KLV2 A0A088ANX5 A0A1L8DH85 A0A293M3K2 A0A2H8TEB9 A0A2S2NIV7 A0A0L7QN64 A0A346KIF7 E2C6F3 A0A0A7W5M9 A0A154PQ81 A0A1L8DHE1 A0A2A3ERA8 A0A3R7SZ08 A0A2S2QX30 J9JQ37 A0A1B0GI91 A0A1L8DH69 A0A1B6IU17 A0A221I039 U5EWF0 T1ITD4 A0A0M4M1U9 A0A1Y1MN18 A0A0K8RHJ2 B7P3R2 A0A131XYX4 A0A1J1IN70 V5GSN9 A0A1W4WTT0 A0A131XHG7 A0A023EMC3 A0A0T6AYF1 Q28ZD6 B4GHX0 B0VZA5 A0A3B0JLR2 A0A182H5G6 A0A224ZAL6 A0A131YPD9 A0A0B4PM30 L7LZN3 J3JXQ7 B4NX50 B4MPI6 G5APN9 A0A182NIF6 B3MD12 A0A1W4YN99 A0A0K8U481 A0A023FHT4 A0A0A9X8N4 A0A182QN88 H3B6Z8 A0A0A1WI47 G3PNU7 A0A3P8XP85 A0A060VZ13
S4PA52 A0A0L7KW91 A0A194QY06 I4DMV0 I4DNB1 A0A3S2NJJ1 A0A067RC76 A0A2P8Y6Z1 A0A2J7QWY3 A0A3L8DJS6 A0A0J7L1Y1 A0A2R7VVD0 V5QNC4 A0A026WMR4 E2AK51 A0A195CU26 A0A195FSM8 A0A158NKX1 F4WAD7 A0A151I2C7 A0A151WMR4 A0A1W7HGV7 A0A195E3E4 A0A0P4VS28 R4G8D4 E9IYZ9 A0A224XWF8 A0A0V0G7D7 R4WCQ7 A0A069DRH1 A0A2R5LK36 A0A1B6DGV8 A0A310SYT0 A0A168GIS3 V9II37 A0A0M8ZYH5 A0A218KLV2 A0A088ANX5 A0A1L8DH85 A0A293M3K2 A0A2H8TEB9 A0A2S2NIV7 A0A0L7QN64 A0A346KIF7 E2C6F3 A0A0A7W5M9 A0A154PQ81 A0A1L8DHE1 A0A2A3ERA8 A0A3R7SZ08 A0A2S2QX30 J9JQ37 A0A1B0GI91 A0A1L8DH69 A0A1B6IU17 A0A221I039 U5EWF0 T1ITD4 A0A0M4M1U9 A0A1Y1MN18 A0A0K8RHJ2 B7P3R2 A0A131XYX4 A0A1J1IN70 V5GSN9 A0A1W4WTT0 A0A131XHG7 A0A023EMC3 A0A0T6AYF1 Q28ZD6 B4GHX0 B0VZA5 A0A3B0JLR2 A0A182H5G6 A0A224ZAL6 A0A131YPD9 A0A0B4PM30 L7LZN3 J3JXQ7 B4NX50 B4MPI6 G5APN9 A0A182NIF6 B3MD12 A0A1W4YN99 A0A0K8U481 A0A023FHT4 A0A0A9X8N4 A0A182QN88 H3B6Z8 A0A0A1WI47 G3PNU7 A0A3P8XP85 A0A060VZ13
Pubmed
19121390
28756777
23622113
26227816
26354079
22651552
+ More
24845553 29403074 30249741 24508170 20798317 21347285 21719571 27129103 21282665 23691247 26334808 27120575 25530093 28004739 28049606 24945155 15632085 17994087 26483478 28797301 26830274 25576852 22516182 17550304 21993625 25401762 26823975 9215903 25830018 25069045 24755649
24845553 29403074 30249741 24508170 20798317 21347285 21719571 27129103 21282665 23691247 26334808 27120575 25530093 28004739 28049606 24945155 15632085 17994087 26483478 28797301 26830274 25576852 22516182 17550304 21993625 25401762 26823975 9215903 25830018 25069045 24755649
EMBL
BABH01028841
BABH01028842
ODYU01000451
SOQ35256.1
KZ150162
PZC72690.1
+ More
JQ731644 AFK64813.1 NWSH01000119 PCG79371.1 GDQN01001825 JAT89229.1 GAIX01003404 JAA89156.1 JTDY01004987 KOB67503.1 KQ460949 KPJ10423.1 AK402618 BAM19240.1 AK402818 KQ459598 BAM19401.1 KPI94542.1 RSAL01000070 RVE49184.1 KK852552 KDR21486.1 PYGN01000853 PSN40015.1 NEVH01009413 PNF33089.1 QOIP01000007 RLU20611.1 LBMM01001227 KMQ96686.1 KK854079 PTY10851.1 KF601293 AHB17949.1 KK107152 EZA57213.1 GL440191 EFN66194.1 KQ977306 KYN03634.1 KQ981281 KYN43302.1 ADTU01018980 GL888048 EGI68820.1 KQ976551 KYM80865.1 KQ982937 KYQ49098.1 LC215245 BAX36489.1 KQ979701 KYN19668.1 GDKW01001354 JAI55241.1 ACPB03011945 GAHY01001167 JAA76343.1 GL767078 EFZ14176.1 GFTR01004125 JAW12301.1 GECL01002108 JAP04016.1 AK417073 BAN20288.1 GBGD01002647 JAC86242.1 GGLE01005766 MBY09892.1 GEDC01012400 JAS24898.1 KQ759770 OAD62937.1 KU363813 ANC33415.1 JR048625 AEY60710.1 KQ435796 KOX73517.1 KX216509 APL97141.1 GFDF01008262 JAV05822.1 GFWV01010773 MAA35502.1 GFXV01000641 MBW12446.1 GGMR01004491 MBY17110.1 KQ414875 KOC59921.1 MF994311 AXP79105.1 GL453132 EFN76477.1 KF999956 KM401573 AIB53746.1 AJB28949.1 KQ435007 KZC13524.1 GFDF01008287 JAV05797.1 KZ288193 PBC34044.1 QCYY01000819 ROT82465.1 GGMS01013092 MBY82295.1 ABLF02033373 AJWK01013575 GFDF01008286 JAV05798.1 GECU01017346 GECU01014778 JAS90360.1 JAS92928.1 KY765909 ASM46959.1 GANO01002977 JAB56894.1 JH431477 KR075823 ALE20542.1 GEZM01028110 JAV86458.1 GADI01003829 JAA69979.1 ABJB010201118 DS630348 EEC01234.1 GEFM01004880 JAP70916.1 CVRI01000056 CRL01667.1 GALX01001277 JAB67189.1 GEFH01002956 JAP65625.1 GAPW01003140 JAC10458.1 LJIG01022525 KRT80113.1 CM000071 EAL25677.2 CH479183 EDW36090.1 DS231813 EDS25707.1 OUUW01000001 SPP74459.1 JXUM01026153 KQ560746 KXJ81028.1 GFPF01012566 MAA23712.1 GEDV01008155 JAP80402.1 KF828769 AIT40211.1 GACK01007779 JAA57255.1 BT128028 AEE62989.1 CM000157 EDW89611.2 CH963849 EDW74025.1 JH166391 EHA98999.1 CH902619 EDV36327.2 GDHF01030795 JAI21519.1 GBBK01003470 JAC21012.1 GBHO01027608 GBRD01006283 GBRD01006281 GDHC01016607 JAG15996.1 JAG59538.1 JAQ02022.1 AXCN02000887 AFYH01045564 GBXI01015991 JAC98300.1 FR904344 CDQ60233.1
JQ731644 AFK64813.1 NWSH01000119 PCG79371.1 GDQN01001825 JAT89229.1 GAIX01003404 JAA89156.1 JTDY01004987 KOB67503.1 KQ460949 KPJ10423.1 AK402618 BAM19240.1 AK402818 KQ459598 BAM19401.1 KPI94542.1 RSAL01000070 RVE49184.1 KK852552 KDR21486.1 PYGN01000853 PSN40015.1 NEVH01009413 PNF33089.1 QOIP01000007 RLU20611.1 LBMM01001227 KMQ96686.1 KK854079 PTY10851.1 KF601293 AHB17949.1 KK107152 EZA57213.1 GL440191 EFN66194.1 KQ977306 KYN03634.1 KQ981281 KYN43302.1 ADTU01018980 GL888048 EGI68820.1 KQ976551 KYM80865.1 KQ982937 KYQ49098.1 LC215245 BAX36489.1 KQ979701 KYN19668.1 GDKW01001354 JAI55241.1 ACPB03011945 GAHY01001167 JAA76343.1 GL767078 EFZ14176.1 GFTR01004125 JAW12301.1 GECL01002108 JAP04016.1 AK417073 BAN20288.1 GBGD01002647 JAC86242.1 GGLE01005766 MBY09892.1 GEDC01012400 JAS24898.1 KQ759770 OAD62937.1 KU363813 ANC33415.1 JR048625 AEY60710.1 KQ435796 KOX73517.1 KX216509 APL97141.1 GFDF01008262 JAV05822.1 GFWV01010773 MAA35502.1 GFXV01000641 MBW12446.1 GGMR01004491 MBY17110.1 KQ414875 KOC59921.1 MF994311 AXP79105.1 GL453132 EFN76477.1 KF999956 KM401573 AIB53746.1 AJB28949.1 KQ435007 KZC13524.1 GFDF01008287 JAV05797.1 KZ288193 PBC34044.1 QCYY01000819 ROT82465.1 GGMS01013092 MBY82295.1 ABLF02033373 AJWK01013575 GFDF01008286 JAV05798.1 GECU01017346 GECU01014778 JAS90360.1 JAS92928.1 KY765909 ASM46959.1 GANO01002977 JAB56894.1 JH431477 KR075823 ALE20542.1 GEZM01028110 JAV86458.1 GADI01003829 JAA69979.1 ABJB010201118 DS630348 EEC01234.1 GEFM01004880 JAP70916.1 CVRI01000056 CRL01667.1 GALX01001277 JAB67189.1 GEFH01002956 JAP65625.1 GAPW01003140 JAC10458.1 LJIG01022525 KRT80113.1 CM000071 EAL25677.2 CH479183 EDW36090.1 DS231813 EDS25707.1 OUUW01000001 SPP74459.1 JXUM01026153 KQ560746 KXJ81028.1 GFPF01012566 MAA23712.1 GEDV01008155 JAP80402.1 KF828769 AIT40211.1 GACK01007779 JAA57255.1 BT128028 AEE62989.1 CM000157 EDW89611.2 CH963849 EDW74025.1 JH166391 EHA98999.1 CH902619 EDV36327.2 GDHF01030795 JAI21519.1 GBBK01003470 JAC21012.1 GBHO01027608 GBRD01006283 GBRD01006281 GDHC01016607 JAG15996.1 JAG59538.1 JAQ02022.1 AXCN02000887 AFYH01045564 GBXI01015991 JAC98300.1 FR904344 CDQ60233.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053240
UP000053268
UP000283053
+ More
UP000027135 UP000245037 UP000235965 UP000279307 UP000036403 UP000053097 UP000000311 UP000078542 UP000078541 UP000005205 UP000007755 UP000078540 UP000075809 UP000078492 UP000015103 UP000053105 UP000005203 UP000053825 UP000008237 UP000076502 UP000242457 UP000283509 UP000007819 UP000092461 UP000001555 UP000183832 UP000192223 UP000001819 UP000008744 UP000002320 UP000268350 UP000069940 UP000249989 UP000002282 UP000007798 UP000006813 UP000075884 UP000007801 UP000192224 UP000075886 UP000008672 UP000007635 UP000265140 UP000193380
UP000027135 UP000245037 UP000235965 UP000279307 UP000036403 UP000053097 UP000000311 UP000078542 UP000078541 UP000005205 UP000007755 UP000078540 UP000075809 UP000078492 UP000015103 UP000053105 UP000005203 UP000053825 UP000008237 UP000076502 UP000242457 UP000283509 UP000007819 UP000092461 UP000001555 UP000183832 UP000192223 UP000001819 UP000008744 UP000002320 UP000268350 UP000069940 UP000249989 UP000002282 UP000007798 UP000006813 UP000075884 UP000007801 UP000192224 UP000075886 UP000008672 UP000007635 UP000265140 UP000193380
PRIDE
Interpro
SUPFAM
SSF47454
SSF47454
ProteinModelPortal
H9J3S7
A0A2H1V3B1
A0A2W1BIX9
I3UIH5
A0A2A4K553
A0A1E1WQD9
+ More
S4PA52 A0A0L7KW91 A0A194QY06 I4DMV0 I4DNB1 A0A3S2NJJ1 A0A067RC76 A0A2P8Y6Z1 A0A2J7QWY3 A0A3L8DJS6 A0A0J7L1Y1 A0A2R7VVD0 V5QNC4 A0A026WMR4 E2AK51 A0A195CU26 A0A195FSM8 A0A158NKX1 F4WAD7 A0A151I2C7 A0A151WMR4 A0A1W7HGV7 A0A195E3E4 A0A0P4VS28 R4G8D4 E9IYZ9 A0A224XWF8 A0A0V0G7D7 R4WCQ7 A0A069DRH1 A0A2R5LK36 A0A1B6DGV8 A0A310SYT0 A0A168GIS3 V9II37 A0A0M8ZYH5 A0A218KLV2 A0A088ANX5 A0A1L8DH85 A0A293M3K2 A0A2H8TEB9 A0A2S2NIV7 A0A0L7QN64 A0A346KIF7 E2C6F3 A0A0A7W5M9 A0A154PQ81 A0A1L8DHE1 A0A2A3ERA8 A0A3R7SZ08 A0A2S2QX30 J9JQ37 A0A1B0GI91 A0A1L8DH69 A0A1B6IU17 A0A221I039 U5EWF0 T1ITD4 A0A0M4M1U9 A0A1Y1MN18 A0A0K8RHJ2 B7P3R2 A0A131XYX4 A0A1J1IN70 V5GSN9 A0A1W4WTT0 A0A131XHG7 A0A023EMC3 A0A0T6AYF1 Q28ZD6 B4GHX0 B0VZA5 A0A3B0JLR2 A0A182H5G6 A0A224ZAL6 A0A131YPD9 A0A0B4PM30 L7LZN3 J3JXQ7 B4NX50 B4MPI6 G5APN9 A0A182NIF6 B3MD12 A0A1W4YN99 A0A0K8U481 A0A023FHT4 A0A0A9X8N4 A0A182QN88 H3B6Z8 A0A0A1WI47 G3PNU7 A0A3P8XP85 A0A060VZ13
S4PA52 A0A0L7KW91 A0A194QY06 I4DMV0 I4DNB1 A0A3S2NJJ1 A0A067RC76 A0A2P8Y6Z1 A0A2J7QWY3 A0A3L8DJS6 A0A0J7L1Y1 A0A2R7VVD0 V5QNC4 A0A026WMR4 E2AK51 A0A195CU26 A0A195FSM8 A0A158NKX1 F4WAD7 A0A151I2C7 A0A151WMR4 A0A1W7HGV7 A0A195E3E4 A0A0P4VS28 R4G8D4 E9IYZ9 A0A224XWF8 A0A0V0G7D7 R4WCQ7 A0A069DRH1 A0A2R5LK36 A0A1B6DGV8 A0A310SYT0 A0A168GIS3 V9II37 A0A0M8ZYH5 A0A218KLV2 A0A088ANX5 A0A1L8DH85 A0A293M3K2 A0A2H8TEB9 A0A2S2NIV7 A0A0L7QN64 A0A346KIF7 E2C6F3 A0A0A7W5M9 A0A154PQ81 A0A1L8DHE1 A0A2A3ERA8 A0A3R7SZ08 A0A2S2QX30 J9JQ37 A0A1B0GI91 A0A1L8DH69 A0A1B6IU17 A0A221I039 U5EWF0 T1ITD4 A0A0M4M1U9 A0A1Y1MN18 A0A0K8RHJ2 B7P3R2 A0A131XYX4 A0A1J1IN70 V5GSN9 A0A1W4WTT0 A0A131XHG7 A0A023EMC3 A0A0T6AYF1 Q28ZD6 B4GHX0 B0VZA5 A0A3B0JLR2 A0A182H5G6 A0A224ZAL6 A0A131YPD9 A0A0B4PM30 L7LZN3 J3JXQ7 B4NX50 B4MPI6 G5APN9 A0A182NIF6 B3MD12 A0A1W4YN99 A0A0K8U481 A0A023FHT4 A0A0A9X8N4 A0A182QN88 H3B6Z8 A0A0A1WI47 G3PNU7 A0A3P8XP85 A0A060VZ13
PDB
5VPF
E-value=6.45228e-14,
Score=185
Ontologies
PATHWAY
04013
MAPK signaling pathway - fly - Bombyx mori (domestic silkworm)
04137 Mitophagy - animal - Bombyx mori (domestic silkworm)
04214 Apoptosis - fly - Bombyx mori (domestic silkworm)
04310 Wnt signaling pathway - Bombyx mori (domestic silkworm)
04624 Toll and Imd signaling pathway - Bombyx mori (domestic silkworm)
04933 AGE-RAGE signaling pathway in diabetic complications - Bombyx mori (domestic silkworm)
04137 Mitophagy - animal - Bombyx mori (domestic silkworm)
04214 Apoptosis - fly - Bombyx mori (domestic silkworm)
04310 Wnt signaling pathway - Bombyx mori (domestic silkworm)
04624 Toll and Imd signaling pathway - Bombyx mori (domestic silkworm)
04933 AGE-RAGE signaling pathway in diabetic complications - Bombyx mori (domestic silkworm)
GO
GO:0005634
GO:0003677
GO:0006357
GO:0003700
GO:0051591
GO:0034097
GO:0000790
GO:0009314
GO:0000978
GO:0003713
GO:0005667
GO:0000122
GO:0008134
GO:0045944
GO:0032870
GO:0042493
GO:0010033
GO:0005654
GO:0032496
GO:0051726
GO:0009612
GO:0000981
GO:0045597
GO:0001938
GO:0007179
GO:0005719
GO:0035026
GO:1990441
GO:0007265
GO:0043923
GO:0061029
GO:1902895
GO:0070412
GO:0043922
GO:0043065
GO:0043392
GO:0070374
GO:0001525
GO:0048146
GO:0031625
GO:0031953
GO:0017053
GO:0035497
GO:0030224
GO:0001889
GO:0071276
GO:0008285
GO:0035976
GO:0003682
GO:0034614
GO:0001228
GO:1904707
GO:0043524
GO:0001102
GO:0031103
GO:0001774
GO:0003151
GO:0000980
GO:0060395
GO:0005096
GO:0046982
GO:0071277
GO:0010634
GO:2000144
GO:0042803
GO:0035994
GO:0046686
GO:0007420
GO:0008013
GO:0051597
GO:0060070
GO:0006355
GO:0043565
GO:0005524
GO:0006139
GO:0019205
GO:0043564
GO:0006418
GO:0003824
PANTHER
Topology
Subcellular location
Nucleus
Length:
239
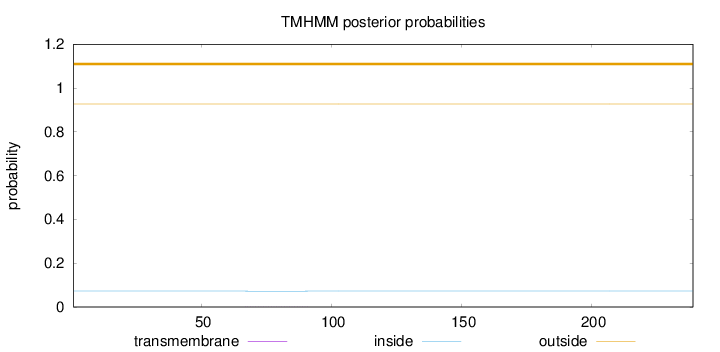
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01277
Exp number, first 60 AAs:
0
Total prob of N-in:
0.07258
outside
1 - 239
Population Genetic Test Statistics
Pi
15.955242
Theta
14.081047
Tajima's D
0.398741
CLR
1.124472
CSRT
0.494275286235688
Interpretation
Uncertain
