Pre Gene Modal
BGIBMGA012627
Annotation
dnaJ_homolog_subfamily_C_member_2_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 3.684
Sequence
CDS
ATGGCCGAGAAAGATTCAAATAGTAATTGTCGTATAGTATCATTAGCGTGGCCATGTGTCAAAAAGAAAGTGGAATGCGTAGGGTCAGCATTCCTCAGATATTATAATATTAAATGTCACGGCGATGATCCATCCTTTACCGAGTCCAAATCTCACGAGAAGGCGGAGGAGATAGTCTTCGATGACGATGTGGAGTACTTGCGAAGCCTCGACCCTAAGGAGTGGAAGTCACAGGATCACTATGCGGTACTTGGTATGAAGGAACTTAGATATGAAGCTACTGATGACGACATTAAACGCGCTTACCGGCAAAAGGTTTTGAAACATCATCCCGACAAGCGTAAGGCGCAGGGCGAGGACATCAGGAGTGACGATGATTACTTTACATGTATCACTAAAGCATATGAAATCCTCGGGACACCAGTCAAACGTAGGTCTTATGATTCAGTTGACCACACTGTGGATGATACCATCCCCTCAACGGCTGAGATTAAGAAAGAAGGATTTTTCGAAGTATTTTCTAAACATTTTGAATCAAATGCTAGGTGGTCAGAAAAAAAGAATGTTCCATTACTTGGTGATGAGAACAGCTCTCGGGAACAGGTAGAAAGGTTTTATGCATTTTGGTATGAGTTTGAATCGTGGAGAGAATTCTCGTATCTTGATGAGGAAGAGAAGGAGAAGGGGGCTGATAGAGAGGAAAGAAGATGGATAGAGAAGCAGAATAAAGCTGCTAGAGCTAAACTAAAGAAAGAAGAGATGGCAAGGTTGAGGAGTTTGGTAGATCTGGCTTATACTTACGATCCTAGAATACAGAGGTTTAAGCAAGAGGACAAGGATAAAAAACTGGCTGCCAAAAAAGCCAGACAGGATGCTGTTCAGGCTAAAAAAGCAGAAGAAGAGCGTATAATGAAGGAAGCTCTCCTCGCTAAGCAAAAAGCAGAAGAGGCAGAGAGAGCACGTCTGGATGCTGCTAGAGCTGAACGAGAAATACAAAAGAAAAATTTACGCAAGGAACGAAAGGCACTTCGTGATCTTTGTAAGGCAAATAACTATTATGCCCAAGGGGAAGATCAAACTGTGACACATATGGCAGCTGTTGAGAAAATCTGTGAAGTCATGAAACTCACTGAACTTCAAGATTTTATGAAAGAACTTGGAATTAATGGTCGAATTGCTTTTGTTAAGACTGTGCAAGAAACAGAAGAAAAACTTGAAGCAGAGAGGAAAGCATTATTTGATACAAAGAAACAAGATGAGCAGAAAATAAAGAAAGATGCTGTTCTGAGGACACCTGTTGAGTGGACTGTTGAGATGACACAGATGCTCATTAAGGCTGTGAATTTATTCCCTGCTGGCACAAACCAGAGGTGGGAAGTGGTGGCAAATTTCTTGAACCAGCATTGTACATTTATTGATGACAAACGGCTATCAGCAAAAGAAGTATTAAACAAAGCTAAGGACTTACAAAGTTCAGACTTTTCAAAAAGTAGTCTAAAGAAAGCTGCCAATGAAGAGGCCTTTGACCAGTTTGAAAAAGAAAAGAAAAAAATTTCCAATCACGTAGACAATACCGGAATTTCAAAGAGCGATAAATTGGTGAATGGAACTACCACTGCTGAGATCAAGCCTGAAGAAAAACCCTGGACCAAAACAGAACAAGAATTGTTGGAACAAGCAATTAAGACGTTTCCAGTAAACACTTCAGAGCGGTGGGAGAAGATCTCTGACTGTATCCCGAATCGATCAAAAAAAGACTGCATGAAGAGATATAAAGAGTTAGTTGAATTAGTAAAAGCAAAGAAACAAGCCGCAAACTTGTCTAAATAA
Protein
MAEKDSNSNCRIVSLAWPCVKKKVECVGSAFLRYYNIKCHGDDPSFTESKSHEKAEEIVFDDDVEYLRSLDPKEWKSQDHYAVLGMKELRYEATDDDIKRAYRQKVLKHHPDKRKAQGEDIRSDDDYFTCITKAYEILGTPVKRRSYDSVDHTVDDTIPSTAEIKKEGFFEVFSKHFESNARWSEKKNVPLLGDENSSREQVERFYAFWYEFESWREFSYLDEEEKEKGADREERRWIEKQNKAARAKLKKEEMARLRSLVDLAYTYDPRIQRFKQEDKDKKLAAKKARQDAVQAKKAEEERIMKEALLAKQKAEEAERARLDAARAEREIQKKNLRKERKALRDLCKANNYYAQGEDQTVTHMAAVEKICEVMKLTELQDFMKELGINGRIAFVKTVQETEEKLEAERKALFDTKKQDEQKIKKDAVLRTPVEWTVEMTQMLIKAVNLFPAGTNQRWEVVANFLNQHCTFIDDKRLSAKEVLNKAKDLQSSDFSKSSLKKAANEEAFDQFEKEKKKISNHVDNTGISKSDKLVNGTTTAEIKPEEKPWTKTEQELLEQAIKTFPVNTSERWEKISDCIPNRSKKDCMKRYKELVELVKAKKQAANLSK
Summary
Similarity
Belongs to the EF-1-beta/EF-1-delta family.
Belongs to the TRAFAC class TrmE-Era-EngA-EngB-Septin-like GTPase superfamily. Septin GTPase family.
Belongs to the TRAFAC class TrmE-Era-EngA-EngB-Septin-like GTPase superfamily. Septin GTPase family.
Uniprot
H6VTR3
H9JSW5
A0A2H1VI51
A0A2A4JTE6
A0A2W1BMD3
A0A0N1I4W4
+ More
A0A0N1IB61 A0A0L7LCC7 A0A3S2L914 A0A1L8DSE0 A0A1B0GJE5 D6WMD9 A0A336MKF7 A0A212F2G4 A0A2A3E660 A0A088AUY0 A0A0M8ZRR3 A0A2P8ZAX1 K7IXT0 A0A0L7QL40 A0A1I8N5J9 T1PGK7 W8BZV8 A0A151I5I7 A0A3L8D807 E2BJ12 A0A026WBB9 A0A232EPA7 A0A158P253 A0A195FH25 E2ARS6 A0A336M9H4 F4WE88 A0A0K8TN36 A0A1Y1KNC8 A0A195D385 E0VGD2 A0A0C9RM22 B4LEC1 B4KWK8 A0A0A1XRW7 A0A0M4EML5 A0A151J503 A0A034WIR4 B4N6I0 A0A0K8VEC3 B4IYW5 A0A0J7KZC6 A0A1I8P0R5 A0A2R7WLU3 N6TTU0 A0A1W4WMD6 B4QJN2 A0A0L0CKI5 A0A1A9VED8 A0A1A9W7I6 A0A1A9Z4L1 A0A1A9XHH9 A0A1B0BHV0 B0WW59 A0A1B0FA59 A0A1Q3EX95 Q7QJJ0 A0A1Q3EX01 B3M6U1 B4PE72 A0A1Q3EWU1 B3NIV8 Q9VP77 B4IAJ8 A0A1J1IT34 A0A1J1ITC7 A0A1B0DID7 E9FZM1 A0A3B0JAU5 Q29E37 B4H401 A0A0A9X5L4 A0A0P4VR15 R4G3D4 A0A0V0G6V0 A0A224XM04 A0A2H8TK11 A0A2S2NPR4 J9JQU3 A0A2S2QGZ4 A0A1D2MCN8 A0A293LS56 A0A1Y9IWD8 T1JNI0 A0A2R5LKW6 A0A3Q3WKQ9 I3JMR1 H3CHP8 A0A2C9GUK5 U3K749 A0A182GGS7 A0A1Z5L4T0 A0A3M0KB53 A0A218UAY1
A0A0N1IB61 A0A0L7LCC7 A0A3S2L914 A0A1L8DSE0 A0A1B0GJE5 D6WMD9 A0A336MKF7 A0A212F2G4 A0A2A3E660 A0A088AUY0 A0A0M8ZRR3 A0A2P8ZAX1 K7IXT0 A0A0L7QL40 A0A1I8N5J9 T1PGK7 W8BZV8 A0A151I5I7 A0A3L8D807 E2BJ12 A0A026WBB9 A0A232EPA7 A0A158P253 A0A195FH25 E2ARS6 A0A336M9H4 F4WE88 A0A0K8TN36 A0A1Y1KNC8 A0A195D385 E0VGD2 A0A0C9RM22 B4LEC1 B4KWK8 A0A0A1XRW7 A0A0M4EML5 A0A151J503 A0A034WIR4 B4N6I0 A0A0K8VEC3 B4IYW5 A0A0J7KZC6 A0A1I8P0R5 A0A2R7WLU3 N6TTU0 A0A1W4WMD6 B4QJN2 A0A0L0CKI5 A0A1A9VED8 A0A1A9W7I6 A0A1A9Z4L1 A0A1A9XHH9 A0A1B0BHV0 B0WW59 A0A1B0FA59 A0A1Q3EX95 Q7QJJ0 A0A1Q3EX01 B3M6U1 B4PE72 A0A1Q3EWU1 B3NIV8 Q9VP77 B4IAJ8 A0A1J1IT34 A0A1J1ITC7 A0A1B0DID7 E9FZM1 A0A3B0JAU5 Q29E37 B4H401 A0A0A9X5L4 A0A0P4VR15 R4G3D4 A0A0V0G6V0 A0A224XM04 A0A2H8TK11 A0A2S2NPR4 J9JQU3 A0A2S2QGZ4 A0A1D2MCN8 A0A293LS56 A0A1Y9IWD8 T1JNI0 A0A2R5LKW6 A0A3Q3WKQ9 I3JMR1 H3CHP8 A0A2C9GUK5 U3K749 A0A182GGS7 A0A1Z5L4T0 A0A3M0KB53 A0A218UAY1
Pubmed
26434795
19121390
28756777
26354079
26227816
18362917
+ More
19820115 22118469 29403074 20075255 25315136 24495485 30249741 20798317 24508170 28648823 21347285 21719571 26369729 28004739 20566863 17994087 25830018 25348373 18057021 23537049 22936249 26108605 12364791 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21292972 15632085 25401762 26823975 27129103 27289101 25186727 15496914 26483478 28528879
19820115 22118469 29403074 20075255 25315136 24495485 30249741 20798317 24508170 28648823 21347285 21719571 26369729 28004739 20566863 17994087 25830018 25348373 18057021 23537049 22936249 26108605 12364791 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21292972 15632085 25401762 26823975 27129103 27289101 25186727 15496914 26483478 28528879
EMBL
JN872898
AFC01237.1
BABH01035997
BABH01035998
BABH01035999
ODYU01002684
+ More
SOQ40503.1 NWSH01000613 PCG75307.1 KZ149965 PZC76219.1 KQ461184 KPJ07530.1 KQ459449 KPJ00878.1 JTDY01001734 KOB73045.1 RSAL01000455 RVE41655.1 GFDF01004824 JAV09260.1 AJWK01020223 AJWK01020224 KQ971343 EFA03316.2 UFQT01001176 SSX29343.1 AGBW02010741 OWR47938.1 KZ288356 PBC27195.1 KQ435883 KOX69775.1 PYGN01000121 PSN53630.1 KQ414934 KOC59320.1 KA647937 AFP62566.1 GAMC01001638 JAC04918.1 KQ976425 KYM88066.1 QOIP01000012 RLU16213.1 GL448543 EFN84357.1 KK107302 EZA52956.1 NNAY01002981 OXU20183.1 ADTU01000084 KQ981606 KYN39683.1 GL442170 EFN63844.1 UFQT01000530 SSX25027.1 GL888102 EGI67526.1 GDAI01002263 JAI15340.1 GEZM01081314 JAV61710.1 KQ976956 KYN06829.1 DS235137 EEB12438.1 GBYB01014432 JAG84199.1 CH940647 EDW70097.1 CH933809 EDW19637.1 GBXI01000642 JAD13650.1 CP012525 ALC43017.1 KQ980071 KYN17859.1 GAKP01004917 JAC54035.1 CH964161 EDW79969.1 KRF99200.1 GDHF01015030 JAI37284.1 CH916366 EDV97673.1 LBMM01001893 KMQ95594.1 KK855054 PTY20576.1 APGK01054900 KB741253 ENN71806.1 CM000363 CM002912 EDX11332.1 KMZ00931.1 JRES01000267 KNC32780.1 JXJN01014603 DS232139 EDS35891.1 CCAG010006049 GFDL01015149 JAV19896.1 AAAB01008807 EAA03986.4 GFDL01015212 JAV19833.1 CH902618 EDV39777.1 CM000159 EDW95085.1 GFDL01015264 JAV19781.1 CH954178 EDV52604.1 AE014296 AY069523 AAF51678.1 AAL39668.1 CH480826 EDW44311.1 CVRI01000059 CRL03367.1 CRL03368.1 AJVK01034355 GL732528 EFX87080.1 OUUW01000002 SPP77122.1 CH379070 EAL30226.2 CH479208 EDW31116.1 GBHO01027597 GBRD01015911 GDHC01007741 JAG16007.1 JAG49915.1 JAQ10888.1 GDKW01001738 JAI54857.1 ACPB03001919 GAHY01002300 JAA75210.1 GECL01002358 JAP03766.1 GFTR01007375 JAW09051.1 GFXV01002692 MBW14497.1 GGMR01006525 MBY19144.1 ABLF02032475 GGMS01007768 MBY76971.1 LJIJ01001804 ODM90758.1 GFWV01006156 MAA30886.1 JH431734 GGLE01006044 MBY10170.1 AERX01052460 AERX01052461 AXCM01004497 AGTO01020852 JXUM01011155 KQ560324 KXJ83037.1 GFJQ02004601 JAW02369.1 QRBI01000112 RMC10368.1 MUZQ01000487 OWK50894.1
SOQ40503.1 NWSH01000613 PCG75307.1 KZ149965 PZC76219.1 KQ461184 KPJ07530.1 KQ459449 KPJ00878.1 JTDY01001734 KOB73045.1 RSAL01000455 RVE41655.1 GFDF01004824 JAV09260.1 AJWK01020223 AJWK01020224 KQ971343 EFA03316.2 UFQT01001176 SSX29343.1 AGBW02010741 OWR47938.1 KZ288356 PBC27195.1 KQ435883 KOX69775.1 PYGN01000121 PSN53630.1 KQ414934 KOC59320.1 KA647937 AFP62566.1 GAMC01001638 JAC04918.1 KQ976425 KYM88066.1 QOIP01000012 RLU16213.1 GL448543 EFN84357.1 KK107302 EZA52956.1 NNAY01002981 OXU20183.1 ADTU01000084 KQ981606 KYN39683.1 GL442170 EFN63844.1 UFQT01000530 SSX25027.1 GL888102 EGI67526.1 GDAI01002263 JAI15340.1 GEZM01081314 JAV61710.1 KQ976956 KYN06829.1 DS235137 EEB12438.1 GBYB01014432 JAG84199.1 CH940647 EDW70097.1 CH933809 EDW19637.1 GBXI01000642 JAD13650.1 CP012525 ALC43017.1 KQ980071 KYN17859.1 GAKP01004917 JAC54035.1 CH964161 EDW79969.1 KRF99200.1 GDHF01015030 JAI37284.1 CH916366 EDV97673.1 LBMM01001893 KMQ95594.1 KK855054 PTY20576.1 APGK01054900 KB741253 ENN71806.1 CM000363 CM002912 EDX11332.1 KMZ00931.1 JRES01000267 KNC32780.1 JXJN01014603 DS232139 EDS35891.1 CCAG010006049 GFDL01015149 JAV19896.1 AAAB01008807 EAA03986.4 GFDL01015212 JAV19833.1 CH902618 EDV39777.1 CM000159 EDW95085.1 GFDL01015264 JAV19781.1 CH954178 EDV52604.1 AE014296 AY069523 AAF51678.1 AAL39668.1 CH480826 EDW44311.1 CVRI01000059 CRL03367.1 CRL03368.1 AJVK01034355 GL732528 EFX87080.1 OUUW01000002 SPP77122.1 CH379070 EAL30226.2 CH479208 EDW31116.1 GBHO01027597 GBRD01015911 GDHC01007741 JAG16007.1 JAG49915.1 JAQ10888.1 GDKW01001738 JAI54857.1 ACPB03001919 GAHY01002300 JAA75210.1 GECL01002358 JAP03766.1 GFTR01007375 JAW09051.1 GFXV01002692 MBW14497.1 GGMR01006525 MBY19144.1 ABLF02032475 GGMS01007768 MBY76971.1 LJIJ01001804 ODM90758.1 GFWV01006156 MAA30886.1 JH431734 GGLE01006044 MBY10170.1 AERX01052460 AERX01052461 AXCM01004497 AGTO01020852 JXUM01011155 KQ560324 KXJ83037.1 GFJQ02004601 JAW02369.1 QRBI01000112 RMC10368.1 MUZQ01000487 OWK50894.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000037510
UP000283053
+ More
UP000092461 UP000007266 UP000007151 UP000242457 UP000005203 UP000053105 UP000245037 UP000002358 UP000053825 UP000095301 UP000078540 UP000279307 UP000008237 UP000053097 UP000215335 UP000005205 UP000078541 UP000000311 UP000007755 UP000078542 UP000009046 UP000008792 UP000009192 UP000092553 UP000078492 UP000007798 UP000001070 UP000036403 UP000095300 UP000019118 UP000192223 UP000000304 UP000037069 UP000078200 UP000091820 UP000092445 UP000092443 UP000092460 UP000002320 UP000092444 UP000007062 UP000007801 UP000002282 UP000008711 UP000000803 UP000001292 UP000183832 UP000092462 UP000000305 UP000268350 UP000001819 UP000008744 UP000015103 UP000007819 UP000094527 UP000075920 UP000261620 UP000005207 UP000007303 UP000075883 UP000016665 UP000069940 UP000249989 UP000269221 UP000197619
UP000092461 UP000007266 UP000007151 UP000242457 UP000005203 UP000053105 UP000245037 UP000002358 UP000053825 UP000095301 UP000078540 UP000279307 UP000008237 UP000053097 UP000215335 UP000005205 UP000078541 UP000000311 UP000007755 UP000078542 UP000009046 UP000008792 UP000009192 UP000092553 UP000078492 UP000007798 UP000001070 UP000036403 UP000095300 UP000019118 UP000192223 UP000000304 UP000037069 UP000078200 UP000091820 UP000092445 UP000092443 UP000092460 UP000002320 UP000092444 UP000007062 UP000007801 UP000002282 UP000008711 UP000000803 UP000001292 UP000183832 UP000092462 UP000000305 UP000268350 UP000001819 UP000008744 UP000015103 UP000007819 UP000094527 UP000075920 UP000261620 UP000005207 UP000007303 UP000075883 UP000016665 UP000069940 UP000249989 UP000269221 UP000197619
Pfam
Interpro
IPR017884
SANT_dom
+ More
IPR009057 Homeobox-like_sf
IPR036869 J_dom_sf
IPR001005 SANT/Myb
IPR017877 Myb-like_dom
IPR032003 RAC_head
IPR001623 DnaJ_domain
IPR017930 Myb_dom
IPR002035 VWF_A
IPR018253 DnaJ_domain_CS
IPR041426 Mos1_HTH
IPR014717 Transl_elong_EF1B/ribosomal_S6
IPR001326 Transl_elong_EF1B_B/D_CS
IPR014038 EF1B_bsu/dsu_GNE
IPR036219 eEF-1beta-like_sf
IPR018940 EF-1_beta_acid_region_euk
IPR036465 vWFA_dom_sf
IPR027417 P-loop_NTPase
IPR030379 G_SEPTIN_dom
IPR016491 Septin
IPR000719 Prot_kinase_dom
IPR011009 Kinase-like_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR009057 Homeobox-like_sf
IPR036869 J_dom_sf
IPR001005 SANT/Myb
IPR017877 Myb-like_dom
IPR032003 RAC_head
IPR001623 DnaJ_domain
IPR017930 Myb_dom
IPR002035 VWF_A
IPR018253 DnaJ_domain_CS
IPR041426 Mos1_HTH
IPR014717 Transl_elong_EF1B/ribosomal_S6
IPR001326 Transl_elong_EF1B_B/D_CS
IPR014038 EF1B_bsu/dsu_GNE
IPR036219 eEF-1beta-like_sf
IPR018940 EF-1_beta_acid_region_euk
IPR036465 vWFA_dom_sf
IPR027417 P-loop_NTPase
IPR030379 G_SEPTIN_dom
IPR016491 Septin
IPR000719 Prot_kinase_dom
IPR011009 Kinase-like_dom_sf
IPR017441 Protein_kinase_ATP_BS
SUPFAM
Gene 3D
ProteinModelPortal
H6VTR3
H9JSW5
A0A2H1VI51
A0A2A4JTE6
A0A2W1BMD3
A0A0N1I4W4
+ More
A0A0N1IB61 A0A0L7LCC7 A0A3S2L914 A0A1L8DSE0 A0A1B0GJE5 D6WMD9 A0A336MKF7 A0A212F2G4 A0A2A3E660 A0A088AUY0 A0A0M8ZRR3 A0A2P8ZAX1 K7IXT0 A0A0L7QL40 A0A1I8N5J9 T1PGK7 W8BZV8 A0A151I5I7 A0A3L8D807 E2BJ12 A0A026WBB9 A0A232EPA7 A0A158P253 A0A195FH25 E2ARS6 A0A336M9H4 F4WE88 A0A0K8TN36 A0A1Y1KNC8 A0A195D385 E0VGD2 A0A0C9RM22 B4LEC1 B4KWK8 A0A0A1XRW7 A0A0M4EML5 A0A151J503 A0A034WIR4 B4N6I0 A0A0K8VEC3 B4IYW5 A0A0J7KZC6 A0A1I8P0R5 A0A2R7WLU3 N6TTU0 A0A1W4WMD6 B4QJN2 A0A0L0CKI5 A0A1A9VED8 A0A1A9W7I6 A0A1A9Z4L1 A0A1A9XHH9 A0A1B0BHV0 B0WW59 A0A1B0FA59 A0A1Q3EX95 Q7QJJ0 A0A1Q3EX01 B3M6U1 B4PE72 A0A1Q3EWU1 B3NIV8 Q9VP77 B4IAJ8 A0A1J1IT34 A0A1J1ITC7 A0A1B0DID7 E9FZM1 A0A3B0JAU5 Q29E37 B4H401 A0A0A9X5L4 A0A0P4VR15 R4G3D4 A0A0V0G6V0 A0A224XM04 A0A2H8TK11 A0A2S2NPR4 J9JQU3 A0A2S2QGZ4 A0A1D2MCN8 A0A293LS56 A0A1Y9IWD8 T1JNI0 A0A2R5LKW6 A0A3Q3WKQ9 I3JMR1 H3CHP8 A0A2C9GUK5 U3K749 A0A182GGS7 A0A1Z5L4T0 A0A3M0KB53 A0A218UAY1
A0A0N1IB61 A0A0L7LCC7 A0A3S2L914 A0A1L8DSE0 A0A1B0GJE5 D6WMD9 A0A336MKF7 A0A212F2G4 A0A2A3E660 A0A088AUY0 A0A0M8ZRR3 A0A2P8ZAX1 K7IXT0 A0A0L7QL40 A0A1I8N5J9 T1PGK7 W8BZV8 A0A151I5I7 A0A3L8D807 E2BJ12 A0A026WBB9 A0A232EPA7 A0A158P253 A0A195FH25 E2ARS6 A0A336M9H4 F4WE88 A0A0K8TN36 A0A1Y1KNC8 A0A195D385 E0VGD2 A0A0C9RM22 B4LEC1 B4KWK8 A0A0A1XRW7 A0A0M4EML5 A0A151J503 A0A034WIR4 B4N6I0 A0A0K8VEC3 B4IYW5 A0A0J7KZC6 A0A1I8P0R5 A0A2R7WLU3 N6TTU0 A0A1W4WMD6 B4QJN2 A0A0L0CKI5 A0A1A9VED8 A0A1A9W7I6 A0A1A9Z4L1 A0A1A9XHH9 A0A1B0BHV0 B0WW59 A0A1B0FA59 A0A1Q3EX95 Q7QJJ0 A0A1Q3EX01 B3M6U1 B4PE72 A0A1Q3EWU1 B3NIV8 Q9VP77 B4IAJ8 A0A1J1IT34 A0A1J1ITC7 A0A1B0DID7 E9FZM1 A0A3B0JAU5 Q29E37 B4H401 A0A0A9X5L4 A0A0P4VR15 R4G3D4 A0A0V0G6V0 A0A224XM04 A0A2H8TK11 A0A2S2NPR4 J9JQU3 A0A2S2QGZ4 A0A1D2MCN8 A0A293LS56 A0A1Y9IWD8 T1JNI0 A0A2R5LKW6 A0A3Q3WKQ9 I3JMR1 H3CHP8 A0A2C9GUK5 U3K749 A0A182GGS7 A0A1Z5L4T0 A0A3M0KB53 A0A218UAY1
PDB
5DJE
E-value=1.53481e-17,
Score=221
Ontologies
GO
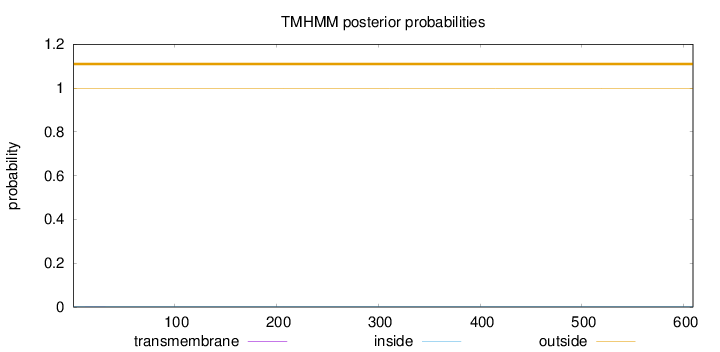
Topology
Subcellular location
Nucleus
Length:
609
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00354
Exp number, first 60 AAs:
0.00349
Total prob of N-in:
0.00241
outside
1 - 609
Population Genetic Test Statistics
Pi
188.867554
Theta
164.102303
Tajima's D
0.503358
CLR
0.08698
CSRT
0.52082395880206
Interpretation
Uncertain
