Gene
KWMTBOMO10681
Pre Gene Modal
BGIBMGA008250
Annotation
PREDICTED:_gastrula_zinc_finger_protein_XlCGF57.1-like_[Amyelois_transitella]
Full name
Zinc finger protein
Transcription factor
Location in the cell
Nuclear Reliability : 4.694
Sequence
CDS
ATGGAGTCCAAAATATGTCGTTTATGTCTCGTTCAACCAGGAAATATTAATTTGTTCGAAACACACGAAGATAATGTGCAATATTGTAAGAAAATTATGCATTGTGCGAACATCCACATCGCAGAGGACGACGGCCTGCCTGATTTCATGTGCGAAGAATGCCTAGTTAAACTTTCTGTTGCCTTCGATTTCATCACCAAATGCCAAGCCTCTGACAAAGCTTTACGTTCAGTATCTTTGAATTATAAACCAGAAAAGAACGACGTCAAATCCGAATACATTGAAATTAAAGAGGAACCTGAAATCAAGTCTGAAACTGAATTCTTAGACGGTGATGATCATCAGTATGAAGATTTACAAGGTAAACCCAGAAAATCATCTAGAACAAAGAGTAGGGGGCCAAGAATCTGTGCAATATGCGGTTACAGGGCTGCCTGTCAGAGTGCCTACAAAGACCATATGAGATCTCATTCTAAAATCAAATTGTTTGAATGTGAAATTTGTCACAAACATTTGTCTGGTAAATATACTCTCAGAAAGCACAAGGAATCGCGCCACGGCATGGAAGTAGTTAGAAAGTTCAAAAAAGAGGCCAGAAATAAAACAATTAATGGTCCAGTACAATGTGTTGTCTGCGGGCAGATACTACCTTCTTCTTCAGCCTTAGAAATTCATAACACAATTCATACAGGAGAGAAACCATATCAATGTGATGTCTGCAATAGATCATTCCGATTAAAGGGTTGTTTGAAAAGGCACATTGAAACTAACCACTTAGAGAGAGAAAAAACATTTATTTGTGAGACATGCGGCAACAGTTTCTACAGGAAAGCAGATATCATAATTCACATTCGTTCTCACACGCAAGAGAAGCCATATGAATGTAAATTATGCCACAGCAAATTCTCCCAACTTTCATCTCTGATACGCCACAAACGAACACATTCTGGAGAGAAAACACATTCCTGCTCAACATGCAACAAACAATTTTATGACAAATATGCTGTGAGGAGACACATGCTGTCACATTCCGATGAGAGGAAACACAAGTGTCACCTTTGTGACAAACTTTTCAAGACGAAATCAGCAAGGAATTTACATCTTTCATTACATACCAATGAGACTCGTTTCATATGCAGCTTCTGTGGAATGATGTTTTCACTTAAAGGTAATCTCAAGACCCATATGATTAGGAAACATTCGGAAAAAGCCGGAGAATGTAAAGTATGTATGAAAGAATTTCCGAATCTAGCTGAACATTTACGCAAACATACCGGAGAAAAACCCTATGTTTGCAGGACCTGCAATCAAGCATTTGCTACACAAAGAAGTCTTTCTTTCCACACCATGTACAAACATGAGAGAGCTGATAAATATCGTTGTACAGTTGGAGAATGTAATAAAGCATTTCCCATGATGAAGTTGCTCGAGACTCATTTGTATAAATATCATACAGACACCATGCCGTTTATGTGCAGTCACTGTCCAAGAAGGTTTTATCGTAGGTCAGATTTGACAAGACACCTTAAAGGTAGTCATTTAGACAGTGACAAACTCAAAGTAAAACCAGAGCTAATATTAACTATAGTGAATATCTAA
Protein
MESKICRLCLVQPGNINLFETHEDNVQYCKKIMHCANIHIAEDDGLPDFMCEECLVKLSVAFDFITKCQASDKALRSVSLNYKPEKNDVKSEYIEIKEEPEIKSETEFLDGDDHQYEDLQGKPRKSSRTKSRGPRICAICGYRAACQSAYKDHMRSHSKIKLFECEICHKHLSGKYTLRKHKESRHGMEVVRKFKKEARNKTINGPVQCVVCGQILPSSSALEIHNTIHTGEKPYQCDVCNRSFRLKGCLKRHIETNHLEREKTFICETCGNSFYRKADIIIHIRSHTQEKPYECKLCHSKFSQLSSLIRHKRTHSGEKTHSCSTCNKQFYDKYAVRRHMLSHSDERKHKCHLCDKLFKTKSARNLHLSLHTNETRFICSFCGMMFSLKGNLKTHMIRKHSEKAGECKVCMKEFPNLAEHLRKHTGEKPYVCRTCNQAFATQRSLSFHTMYKHERADKYRCTVGECNKAFPMMKLLETHLYKYHTDTMPFMCSHCPRRFYRRSDLTRHLKGSHLDSDKLKVKPELILTIVNI
Summary
Uniprot
H9JFF3
A0A2W1BAG8
A0A0L7LJU2
A0A0L7LK64
H9JFG8
A0A3B3C4I9
+ More
A0A3Q3FED3 A0A146XCF7 A0A147AF84 A0A3Q2Z8V3 A0A146P0B7 A0A3Q4AR89 J9JMS6 A0A146TB62 A0A146QI62 A0A3B3Z3J6 A0A146PSZ5 A0A3B3W1H6 A0A087U2Y4 A0A3Q2CMN1 A0A087XHT8 A0A3B4X3Z3 A0A146MKP3 A0A2P6KF50 A0A0F8AD47 V4A9F3 A0A3Q2ZDK0 A0A3Q2NUF1 I3JLU3 A0A3Q0R6D2 J9JJ22 A0A3Q1AXL3 A0A3Q3MBN0 A0A3P9HTS6 X1WP87 A0A3Q3X2B3 A0A3Q2YV76 A0A3Q1BJS5 A0A087YLI0 X1WW20 A0A2P6K297 A0A3Q4ASE0 A0A3Q0SCP5 A0A3B5LBX5 A0A287DEH8 A0A147AAK4 A0A146ZJP7 A0A3P8R268 A0A3B4U5G4 A0A146YZF7 A0A3P9B4C5 A0A3Q1ERZ1 A0A3Q0SJN5 A0A3P8RAE2 A0A3B4U132 A0A3Q2Z783 A0A087UHT5 A0A3Q2Y222 A0A3B4WU99 A0A3P8R2L0 A0A3P8S5G1 A0A146RQL3 A0A3Q1BX91 A0A3B4TXC2 A0A315VYA0 A0A2P6K572 A0A087TDG0 A0A2P6KPN3 A0A2P6K9Y8 A0A2P6K1M6 A0A0L8HUJ2 A0A3Q2YXI6 A0A3B4TZ98 A0A3Q1FB33 C3ZDM5 A0A3P8SJL2 A0A2P6K5F6 R7V818 G3NLR6 A0A3B3HP97 A0A0L8HVP4 A0A3Q2CBV3 A0A3P9B4A9 C3ZPQ0 D3ZIN6 A0A087U3D4 A0A087V162 C3ZUZ5
A0A3Q3FED3 A0A146XCF7 A0A147AF84 A0A3Q2Z8V3 A0A146P0B7 A0A3Q4AR89 J9JMS6 A0A146TB62 A0A146QI62 A0A3B3Z3J6 A0A146PSZ5 A0A3B3W1H6 A0A087U2Y4 A0A3Q2CMN1 A0A087XHT8 A0A3B4X3Z3 A0A146MKP3 A0A2P6KF50 A0A0F8AD47 V4A9F3 A0A3Q2ZDK0 A0A3Q2NUF1 I3JLU3 A0A3Q0R6D2 J9JJ22 A0A3Q1AXL3 A0A3Q3MBN0 A0A3P9HTS6 X1WP87 A0A3Q3X2B3 A0A3Q2YV76 A0A3Q1BJS5 A0A087YLI0 X1WW20 A0A2P6K297 A0A3Q4ASE0 A0A3Q0SCP5 A0A3B5LBX5 A0A287DEH8 A0A147AAK4 A0A146ZJP7 A0A3P8R268 A0A3B4U5G4 A0A146YZF7 A0A3P9B4C5 A0A3Q1ERZ1 A0A3Q0SJN5 A0A3P8RAE2 A0A3B4U132 A0A3Q2Z783 A0A087UHT5 A0A3Q2Y222 A0A3B4WU99 A0A3P8R2L0 A0A3P8S5G1 A0A146RQL3 A0A3Q1BX91 A0A3B4TXC2 A0A315VYA0 A0A2P6K572 A0A087TDG0 A0A2P6KPN3 A0A2P6K9Y8 A0A2P6K1M6 A0A0L8HUJ2 A0A3Q2YXI6 A0A3B4TZ98 A0A3Q1FB33 C3ZDM5 A0A3P8SJL2 A0A2P6K5F6 R7V818 G3NLR6 A0A3B3HP97 A0A0L8HVP4 A0A3Q2CBV3 A0A3P9B4A9 C3ZPQ0 D3ZIN6 A0A087U3D4 A0A087V162 C3ZUZ5
EC Number
2.1.1.-
Pubmed
EMBL
BABH01034947
KZ150415
PZC70964.1
JTDY01000800
KOB75833.1
KOB75837.1
+ More
GCES01046570 JAR39753.1 GCES01009232 JAR77091.1 GCES01149080 JAQ37242.1 ABLF02038601 GCES01096365 JAQ89957.1 GCES01130511 JAQ55811.1 GCES01139239 JAQ47083.1 KK117904 KFM71723.1 AYCK01023677 AYCK01023678 AYCK01023679 AYCK01023680 AYCK01023681 AYCK01023682 AYCK01023683 GCES01166730 JAQ19592.1 MWRG01013681 PRD24960.1 KQ043214 KKF09316.1 KB200701 ESP00624.1 AERX01055903 AERX01055904 AERX01055905 AERX01055906 AERX01055907 ABLF02033404 ABLF02029797 AYCK01001272 AYCK01001273 ABLF02029788 MWRG01038200 PRD20450.1 AGTP01019276 AGTP01019277 GCES01011078 JAR75245.1 GCES01019793 JAR66530.1 GCES01026855 JAR59468.1 KK119857 KFM76924.1 GCES01115895 JAQ70427.1 NHOQ01000797 PWA28608.1 MWRG01026172 PRD21467.1 KK114724 KFM63149.1 MWRG01007387 PRD28287.1 MWRG01018834 PRD23141.1 MWRG01042629 PRD20213.1 KQ417280 KOF92864.1 GG666612 EEN48831.1 MWRG01025837 PRD21530.1 AMQN01038589 KB294404 ELU14642.1 KOF92865.1 GG666659 EEN45471.1 AABR07002870 AABR07002896 KK117968 KFM71873.1 KL818556 KFM83351.1 GG666686 EEN43642.1
GCES01046570 JAR39753.1 GCES01009232 JAR77091.1 GCES01149080 JAQ37242.1 ABLF02038601 GCES01096365 JAQ89957.1 GCES01130511 JAQ55811.1 GCES01139239 JAQ47083.1 KK117904 KFM71723.1 AYCK01023677 AYCK01023678 AYCK01023679 AYCK01023680 AYCK01023681 AYCK01023682 AYCK01023683 GCES01166730 JAQ19592.1 MWRG01013681 PRD24960.1 KQ043214 KKF09316.1 KB200701 ESP00624.1 AERX01055903 AERX01055904 AERX01055905 AERX01055906 AERX01055907 ABLF02033404 ABLF02029797 AYCK01001272 AYCK01001273 ABLF02029788 MWRG01038200 PRD20450.1 AGTP01019276 AGTP01019277 GCES01011078 JAR75245.1 GCES01019793 JAR66530.1 GCES01026855 JAR59468.1 KK119857 KFM76924.1 GCES01115895 JAQ70427.1 NHOQ01000797 PWA28608.1 MWRG01026172 PRD21467.1 KK114724 KFM63149.1 MWRG01007387 PRD28287.1 MWRG01018834 PRD23141.1 MWRG01042629 PRD20213.1 KQ417280 KOF92864.1 GG666612 EEN48831.1 MWRG01025837 PRD21530.1 AMQN01038589 KB294404 ELU14642.1 KOF92865.1 GG666659 EEN45471.1 AABR07002870 AABR07002896 KK117968 KFM71873.1 KL818556 KFM83351.1 GG666686 EEN43642.1
Proteomes
UP000005204
UP000037510
UP000261560
UP000261660
UP000264820
UP000261620
+ More
UP000007819 UP000261480 UP000261500 UP000054359 UP000265020 UP000028760 UP000261360 UP000030746 UP000265000 UP000005207 UP000261340 UP000257160 UP000265200 UP000261380 UP000005215 UP000265100 UP000261420 UP000265160 UP000257200 UP000265080 UP000053454 UP000001554 UP000014760 UP000007635 UP000001038 UP000002494
UP000007819 UP000261480 UP000261500 UP000054359 UP000265020 UP000028760 UP000261360 UP000030746 UP000265000 UP000005207 UP000261340 UP000257160 UP000265200 UP000261380 UP000005215 UP000265100 UP000261420 UP000265160 UP000257200 UP000265080 UP000053454 UP000001554 UP000014760 UP000007635 UP000001038 UP000002494
Interpro
ProteinModelPortal
H9JFF3
A0A2W1BAG8
A0A0L7LJU2
A0A0L7LK64
H9JFG8
A0A3B3C4I9
+ More
A0A3Q3FED3 A0A146XCF7 A0A147AF84 A0A3Q2Z8V3 A0A146P0B7 A0A3Q4AR89 J9JMS6 A0A146TB62 A0A146QI62 A0A3B3Z3J6 A0A146PSZ5 A0A3B3W1H6 A0A087U2Y4 A0A3Q2CMN1 A0A087XHT8 A0A3B4X3Z3 A0A146MKP3 A0A2P6KF50 A0A0F8AD47 V4A9F3 A0A3Q2ZDK0 A0A3Q2NUF1 I3JLU3 A0A3Q0R6D2 J9JJ22 A0A3Q1AXL3 A0A3Q3MBN0 A0A3P9HTS6 X1WP87 A0A3Q3X2B3 A0A3Q2YV76 A0A3Q1BJS5 A0A087YLI0 X1WW20 A0A2P6K297 A0A3Q4ASE0 A0A3Q0SCP5 A0A3B5LBX5 A0A287DEH8 A0A147AAK4 A0A146ZJP7 A0A3P8R268 A0A3B4U5G4 A0A146YZF7 A0A3P9B4C5 A0A3Q1ERZ1 A0A3Q0SJN5 A0A3P8RAE2 A0A3B4U132 A0A3Q2Z783 A0A087UHT5 A0A3Q2Y222 A0A3B4WU99 A0A3P8R2L0 A0A3P8S5G1 A0A146RQL3 A0A3Q1BX91 A0A3B4TXC2 A0A315VYA0 A0A2P6K572 A0A087TDG0 A0A2P6KPN3 A0A2P6K9Y8 A0A2P6K1M6 A0A0L8HUJ2 A0A3Q2YXI6 A0A3B4TZ98 A0A3Q1FB33 C3ZDM5 A0A3P8SJL2 A0A2P6K5F6 R7V818 G3NLR6 A0A3B3HP97 A0A0L8HVP4 A0A3Q2CBV3 A0A3P9B4A9 C3ZPQ0 D3ZIN6 A0A087U3D4 A0A087V162 C3ZUZ5
A0A3Q3FED3 A0A146XCF7 A0A147AF84 A0A3Q2Z8V3 A0A146P0B7 A0A3Q4AR89 J9JMS6 A0A146TB62 A0A146QI62 A0A3B3Z3J6 A0A146PSZ5 A0A3B3W1H6 A0A087U2Y4 A0A3Q2CMN1 A0A087XHT8 A0A3B4X3Z3 A0A146MKP3 A0A2P6KF50 A0A0F8AD47 V4A9F3 A0A3Q2ZDK0 A0A3Q2NUF1 I3JLU3 A0A3Q0R6D2 J9JJ22 A0A3Q1AXL3 A0A3Q3MBN0 A0A3P9HTS6 X1WP87 A0A3Q3X2B3 A0A3Q2YV76 A0A3Q1BJS5 A0A087YLI0 X1WW20 A0A2P6K297 A0A3Q4ASE0 A0A3Q0SCP5 A0A3B5LBX5 A0A287DEH8 A0A147AAK4 A0A146ZJP7 A0A3P8R268 A0A3B4U5G4 A0A146YZF7 A0A3P9B4C5 A0A3Q1ERZ1 A0A3Q0SJN5 A0A3P8RAE2 A0A3B4U132 A0A3Q2Z783 A0A087UHT5 A0A3Q2Y222 A0A3B4WU99 A0A3P8R2L0 A0A3P8S5G1 A0A146RQL3 A0A3Q1BX91 A0A3B4TXC2 A0A315VYA0 A0A2P6K572 A0A087TDG0 A0A2P6KPN3 A0A2P6K9Y8 A0A2P6K1M6 A0A0L8HUJ2 A0A3Q2YXI6 A0A3B4TZ98 A0A3Q1FB33 C3ZDM5 A0A3P8SJL2 A0A2P6K5F6 R7V818 G3NLR6 A0A3B3HP97 A0A0L8HVP4 A0A3Q2CBV3 A0A3P9B4A9 C3ZPQ0 D3ZIN6 A0A087U3D4 A0A087V162 C3ZUZ5
PDB
5V3M
E-value=2.82763e-36,
Score=382
Ontologies
GO
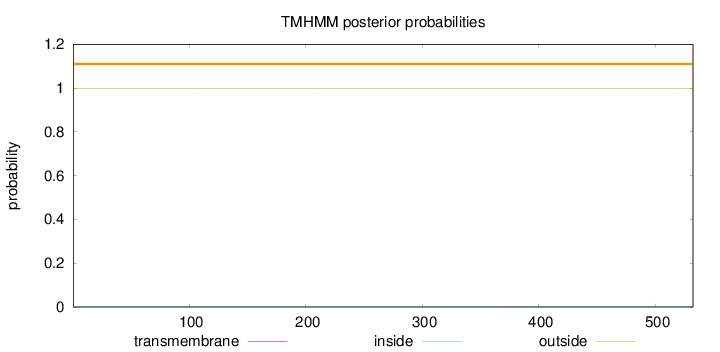
Topology
Length:
532
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00757999999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00183
outside
1 - 532
Population Genetic Test Statistics
Pi
275.995493
Theta
230.673718
Tajima's D
0.573969
CLR
0
CSRT
0.538073096345183
Interpretation
Uncertain
