Gene
KWMTBOMO10472
Pre Gene Modal
BGIBMGA006998
Annotation
PREDICTED:_non-histone_protein_10_[Papilio_polytes]
Transcription factor
Location in the cell
Nuclear Reliability : 4.052
Sequence
CDS
ATGACAGTACAACAGCTCAACTCAAATAATTCTGACGACTCAGATGATATGGTTAGTTTACCGCCCGAAGAAGAAATTTATAGACGAAAATATCAACTGTTATTGGAACGATGTGAAGTCTTGCAACAGGATAACGAAAGAATCATAAACAGGATACACGAAGTGAAAAAAGTGACTAAAAGGTACCGTAAAGATATTAAGTTAGTGATTGAAAGGTTGGACAGACATGGCGATCCTTTTAGGAGCGCTGTATTGGAAATGGATACGAAATCTGAGACTGTCCAGAGACCTGCTCGTCCTGTAGCGAAAGTACAACCAGTCGCCAAACAAACCGACAAACAAAACGGAGGGAAAAAAACAGGAGCCAAACGAAAAACAAAGGCAGAGAAGCCTGAAAAAGATCCTAATGCTCCAAAGAAACCTTGTAATGCATTTTTCCAATTTTGTCAAGAACAGAGGCCTTTGGTAGTTGCTGAAGCTAATTCAGAAGTAGGTGCAGAACCATCCAAACAAGAAGTTACTAGACAATTGGCATCAAGATGGAGAGCTTTGACGAATGATGAAAAAAGGGTATATGTGGCAATGTTTGAGAGGTCAAAAGAGAAATATGCAGAAGAAATGACAGCATATAATATCAAAAAGGAACAATGA
Protein
MTVQQLNSNNSDDSDDMVSLPPEEEIYRRKYQLLLERCEVLQQDNERIINRIHEVKKVTKRYRKDIKLVIERLDRHGDPFRSAVLEMDTKSETVQRPARPVAKVQPVAKQTDKQNGGKKTGAKRKTKAEKPEKDPNAPKKPCNAFFQFCQEQRPLVVAEANSEVGAEPSKQEVTRQLASRWRALTNDEKRVYVAMFERSKEKYAEEMTAYNIKKEQ
Summary
Uniprot
H9JBV3
A0A2H1VIS9
A0A3S2P3R6
S4PFU7
A0A2A4IU83
A0A212ETX9
+ More
A0A194PJ67 A0A194QV30 A0A0L7K551 A0A1B6CCF6 A0A154PJW9 E9JCN2 A0A151K107 A0A151JP39 F4X1R2 A0A026WN12 A0A151X9T2 A0A195B576 A0A158NV75 A0A0J7NT70 A0A195CUM7 A0A2A3ENH5 A0A087ZZI6 E2AFR0 A0A0L7QND5 A0A067QJQ9 A0A232EZR0 A0A1B6KVW4 A0A1B6J6F2 A0A1W4W493 J9KAZ0 E2BF52 C4WW64 A0A3Q0IYF5 T1IES5 A0A0T6B501 A0A2R7X7J7 A0A1L8DWF4 A0A1L8DWD2 A0A1Y1KID1 D6WVJ6 A0A1B0GLZ0 A0A0M9A7B3 A0A023F165 A0A0N7ZA69 A0A2J7QWQ2 A0A2P8XH09 N6U5T0 A0A0B6ZNB2 A0A3S1HKJ3 A7S799 A0A310S4D2 A0A2T7P9B6 K1RBB5 R7VGT2 A0A0B6ZPQ2 A0A3M6TJE0 A0A1S3HRP1 A0A210PHC2 V4AMX9 A0A369RTD4 C3Y9J6 A0A2G8LJP4 A0A163M2R0 A0A1Y1VF09 A0A1Y1WV80 A0A1Y1Z550 A0A163JG73 A0A0L8GHB3 A0A077X0W7 A0A0B7N8W7 A0A1Y1WMB0 A0A168KBZ9 A0A168LBQ6 A0A2G4SU54 A0A1X0RDD0 A0A0C7CJL7 A0A0C7B4N7 A0A367K5X5 A0A068SFV7 A0A1C7MUK1
A0A194PJ67 A0A194QV30 A0A0L7K551 A0A1B6CCF6 A0A154PJW9 E9JCN2 A0A151K107 A0A151JP39 F4X1R2 A0A026WN12 A0A151X9T2 A0A195B576 A0A158NV75 A0A0J7NT70 A0A195CUM7 A0A2A3ENH5 A0A087ZZI6 E2AFR0 A0A0L7QND5 A0A067QJQ9 A0A232EZR0 A0A1B6KVW4 A0A1B6J6F2 A0A1W4W493 J9KAZ0 E2BF52 C4WW64 A0A3Q0IYF5 T1IES5 A0A0T6B501 A0A2R7X7J7 A0A1L8DWF4 A0A1L8DWD2 A0A1Y1KID1 D6WVJ6 A0A1B0GLZ0 A0A0M9A7B3 A0A023F165 A0A0N7ZA69 A0A2J7QWQ2 A0A2P8XH09 N6U5T0 A0A0B6ZNB2 A0A3S1HKJ3 A7S799 A0A310S4D2 A0A2T7P9B6 K1RBB5 R7VGT2 A0A0B6ZPQ2 A0A3M6TJE0 A0A1S3HRP1 A0A210PHC2 V4AMX9 A0A369RTD4 C3Y9J6 A0A2G8LJP4 A0A163M2R0 A0A1Y1VF09 A0A1Y1WV80 A0A1Y1Z550 A0A163JG73 A0A0L8GHB3 A0A077X0W7 A0A0B7N8W7 A0A1Y1WMB0 A0A168KBZ9 A0A168LBQ6 A0A2G4SU54 A0A1X0RDD0 A0A0C7CJL7 A0A0C7B4N7 A0A367K5X5 A0A068SFV7 A0A1C7MUK1
Pubmed
EMBL
BABH01014932
ODYU01002794
SOQ40733.1
RSAL01000031
RVE51618.1
GAIX01003877
+ More
JAA88683.1 NWSH01006781 PCG63239.1 AGBW02012517 OWR44950.1 KQ459603 KPI93083.1 KQ461137 KPJ08835.1 JTDY01010417 KOB58215.1 GEDC01026161 JAS11137.1 KQ434924 KZC11588.1 GL771849 EFZ09436.1 KQ981189 KYN45240.1 KQ978820 KYN28148.1 GL888549 EGI59608.1 KK107167 QOIP01000010 EZA56494.1 RLU17788.1 KQ982365 KYQ57145.1 KQ976595 KYM79641.1 ADTU01026937 LBMM01001888 KMQ95600.1 KQ977279 KYN04388.1 KZ288212 PBC32802.1 GL439124 EFN67739.1 KQ414855 KOC60128.1 KK853278 KDR09064.1 NNAY01001464 OXU23882.1 GEBQ01024381 JAT15596.1 GECU01012960 JAS94746.1 ABLF02028301 GL447936 EFN85678.1 AK341878 BAH72134.1 ACPB03015821 LJIG01009769 KRT82418.1 KK858512 PTY27774.1 GFDF01003322 JAV10762.1 GFDF01003323 JAV10761.1 GEZM01085157 JAV60171.1 KQ971357 EFA08580.1 AJVK01009183 KQ435732 KOX77339.1 GBBI01003978 JAC14734.1 GDRN01103797 GDRN01103796 JAI58016.1 NEVH01009422 PNF33017.1 PYGN01002160 PSN31290.1 APGK01047774 KB741089 KB632382 ENN73937.1 ERL94168.1 HACG01023148 CEK70013.1 RQTK01000339 RUS81396.1 DS469591 EDO40495.1 KQ779356 OAD51995.1 PZQS01000005 PVD30008.1 JH818012 EKC38520.1 AMQN01003942 KB292298 ELU17814.1 HACG01023754 CEK70619.1 RCHS01003480 RMX41542.1 NEDP02076700 OWF35880.1 KB201248 ESO98507.1 NOWV01000238 RDD37502.1 GG666492 EEN63252.1 MRZV01000055 PIK60474.1 LT553414 SAM00728.1 MCFH01000012 ORX53782.1 MCFG01000249 ORX77413.1 MCFE01000026 ORY05336.1 LT552482 SAL99132.1 KQ421836 KOF76249.1 LK023368 CDS13129.1 LN731777 CEP14915.1 MCFD01000001 ORX74701.1 AMYB01000005 OAD02229.1 AMYB01000004 OAD03337.1 KZ303851 PHZ11926.1 KV921869 ORE10063.1 CCYT01000555 CEG83709.1 CCYT01000004 CEG65942.1 PJQL01000266 RCH97565.1 CBTN010000083 CDH60131.1 LUGH01002060 OBZ80490.1
JAA88683.1 NWSH01006781 PCG63239.1 AGBW02012517 OWR44950.1 KQ459603 KPI93083.1 KQ461137 KPJ08835.1 JTDY01010417 KOB58215.1 GEDC01026161 JAS11137.1 KQ434924 KZC11588.1 GL771849 EFZ09436.1 KQ981189 KYN45240.1 KQ978820 KYN28148.1 GL888549 EGI59608.1 KK107167 QOIP01000010 EZA56494.1 RLU17788.1 KQ982365 KYQ57145.1 KQ976595 KYM79641.1 ADTU01026937 LBMM01001888 KMQ95600.1 KQ977279 KYN04388.1 KZ288212 PBC32802.1 GL439124 EFN67739.1 KQ414855 KOC60128.1 KK853278 KDR09064.1 NNAY01001464 OXU23882.1 GEBQ01024381 JAT15596.1 GECU01012960 JAS94746.1 ABLF02028301 GL447936 EFN85678.1 AK341878 BAH72134.1 ACPB03015821 LJIG01009769 KRT82418.1 KK858512 PTY27774.1 GFDF01003322 JAV10762.1 GFDF01003323 JAV10761.1 GEZM01085157 JAV60171.1 KQ971357 EFA08580.1 AJVK01009183 KQ435732 KOX77339.1 GBBI01003978 JAC14734.1 GDRN01103797 GDRN01103796 JAI58016.1 NEVH01009422 PNF33017.1 PYGN01002160 PSN31290.1 APGK01047774 KB741089 KB632382 ENN73937.1 ERL94168.1 HACG01023148 CEK70013.1 RQTK01000339 RUS81396.1 DS469591 EDO40495.1 KQ779356 OAD51995.1 PZQS01000005 PVD30008.1 JH818012 EKC38520.1 AMQN01003942 KB292298 ELU17814.1 HACG01023754 CEK70619.1 RCHS01003480 RMX41542.1 NEDP02076700 OWF35880.1 KB201248 ESO98507.1 NOWV01000238 RDD37502.1 GG666492 EEN63252.1 MRZV01000055 PIK60474.1 LT553414 SAM00728.1 MCFH01000012 ORX53782.1 MCFG01000249 ORX77413.1 MCFE01000026 ORY05336.1 LT552482 SAL99132.1 KQ421836 KOF76249.1 LK023368 CDS13129.1 LN731777 CEP14915.1 MCFD01000001 ORX74701.1 AMYB01000005 OAD02229.1 AMYB01000004 OAD03337.1 KZ303851 PHZ11926.1 KV921869 ORE10063.1 CCYT01000555 CEG83709.1 CCYT01000004 CEG65942.1 PJQL01000266 RCH97565.1 CBTN010000083 CDH60131.1 LUGH01002060 OBZ80490.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000007151
UP000053268
UP000053240
+ More
UP000037510 UP000076502 UP000078541 UP000078492 UP000007755 UP000053097 UP000279307 UP000075809 UP000078540 UP000005205 UP000036403 UP000078542 UP000242457 UP000005203 UP000000311 UP000053825 UP000027135 UP000215335 UP000192223 UP000007819 UP000008237 UP000079169 UP000015103 UP000007266 UP000092462 UP000053105 UP000235965 UP000245037 UP000019118 UP000030742 UP000271974 UP000001593 UP000245119 UP000005408 UP000014760 UP000275408 UP000085678 UP000242188 UP000030746 UP000253843 UP000001554 UP000230750 UP000078561 UP000193719 UP000193944 UP000193498 UP000053454 UP000054107 UP000193922 UP000077051 UP000242254 UP000242414 UP000054919 UP000252139 UP000027586 UP000093000
UP000037510 UP000076502 UP000078541 UP000078492 UP000007755 UP000053097 UP000279307 UP000075809 UP000078540 UP000005205 UP000036403 UP000078542 UP000242457 UP000005203 UP000000311 UP000053825 UP000027135 UP000215335 UP000192223 UP000007819 UP000008237 UP000079169 UP000015103 UP000007266 UP000092462 UP000053105 UP000235965 UP000245037 UP000019118 UP000030742 UP000271974 UP000001593 UP000245119 UP000005408 UP000014760 UP000275408 UP000085678 UP000242188 UP000030746 UP000253843 UP000001554 UP000230750 UP000078561 UP000193719 UP000193944 UP000193498 UP000053454 UP000054107 UP000193922 UP000077051 UP000242254 UP000242414 UP000054919 UP000252139 UP000027586 UP000093000
PRIDE
Pfam
PF00505 HMG_box
SUPFAM
SSF47095
SSF47095
Gene 3D
ProteinModelPortal
H9JBV3
A0A2H1VIS9
A0A3S2P3R6
S4PFU7
A0A2A4IU83
A0A212ETX9
+ More
A0A194PJ67 A0A194QV30 A0A0L7K551 A0A1B6CCF6 A0A154PJW9 E9JCN2 A0A151K107 A0A151JP39 F4X1R2 A0A026WN12 A0A151X9T2 A0A195B576 A0A158NV75 A0A0J7NT70 A0A195CUM7 A0A2A3ENH5 A0A087ZZI6 E2AFR0 A0A0L7QND5 A0A067QJQ9 A0A232EZR0 A0A1B6KVW4 A0A1B6J6F2 A0A1W4W493 J9KAZ0 E2BF52 C4WW64 A0A3Q0IYF5 T1IES5 A0A0T6B501 A0A2R7X7J7 A0A1L8DWF4 A0A1L8DWD2 A0A1Y1KID1 D6WVJ6 A0A1B0GLZ0 A0A0M9A7B3 A0A023F165 A0A0N7ZA69 A0A2J7QWQ2 A0A2P8XH09 N6U5T0 A0A0B6ZNB2 A0A3S1HKJ3 A7S799 A0A310S4D2 A0A2T7P9B6 K1RBB5 R7VGT2 A0A0B6ZPQ2 A0A3M6TJE0 A0A1S3HRP1 A0A210PHC2 V4AMX9 A0A369RTD4 C3Y9J6 A0A2G8LJP4 A0A163M2R0 A0A1Y1VF09 A0A1Y1WV80 A0A1Y1Z550 A0A163JG73 A0A0L8GHB3 A0A077X0W7 A0A0B7N8W7 A0A1Y1WMB0 A0A168KBZ9 A0A168LBQ6 A0A2G4SU54 A0A1X0RDD0 A0A0C7CJL7 A0A0C7B4N7 A0A367K5X5 A0A068SFV7 A0A1C7MUK1
A0A194PJ67 A0A194QV30 A0A0L7K551 A0A1B6CCF6 A0A154PJW9 E9JCN2 A0A151K107 A0A151JP39 F4X1R2 A0A026WN12 A0A151X9T2 A0A195B576 A0A158NV75 A0A0J7NT70 A0A195CUM7 A0A2A3ENH5 A0A087ZZI6 E2AFR0 A0A0L7QND5 A0A067QJQ9 A0A232EZR0 A0A1B6KVW4 A0A1B6J6F2 A0A1W4W493 J9KAZ0 E2BF52 C4WW64 A0A3Q0IYF5 T1IES5 A0A0T6B501 A0A2R7X7J7 A0A1L8DWF4 A0A1L8DWD2 A0A1Y1KID1 D6WVJ6 A0A1B0GLZ0 A0A0M9A7B3 A0A023F165 A0A0N7ZA69 A0A2J7QWQ2 A0A2P8XH09 N6U5T0 A0A0B6ZNB2 A0A3S1HKJ3 A7S799 A0A310S4D2 A0A2T7P9B6 K1RBB5 R7VGT2 A0A0B6ZPQ2 A0A3M6TJE0 A0A1S3HRP1 A0A210PHC2 V4AMX9 A0A369RTD4 C3Y9J6 A0A2G8LJP4 A0A163M2R0 A0A1Y1VF09 A0A1Y1WV80 A0A1Y1Z550 A0A163JG73 A0A0L8GHB3 A0A077X0W7 A0A0B7N8W7 A0A1Y1WMB0 A0A168KBZ9 A0A168LBQ6 A0A2G4SU54 A0A1X0RDD0 A0A0C7CJL7 A0A0C7B4N7 A0A367K5X5 A0A068SFV7 A0A1C7MUK1
PDB
1HMF
E-value=8.05573e-06,
Score=115
Ontologies
PANTHER
Topology
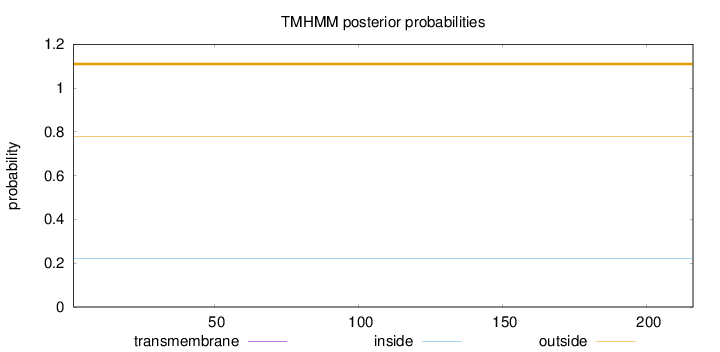
Length:
216
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.22267
outside
1 - 216
Population Genetic Test Statistics
Pi
47.881286
Theta
47.791782
Tajima's D
0.005822
CLR
0.629867
CSRT
0.369881505924704
Interpretation
Uncertain
