Pre Gene Modal
BGIBMGA005634
Annotation
PREDICTED:_protein_shuttle_craft_[Bombyx_mori]
Full name
Protein shuttle craft
Transcription factor
Location in the cell
Extracellular Reliability : 2.664
Sequence
CDS
ATGTGCAGCAGCAAGCTACCGCAAGTGTGCGGGCGTCCTTGCCAACGCATGCTACAGTGCGGCGTACACAAATGCATGAAGGAATGCCACGAGGGTCCCTGTGAAGACTGCACCGAGTCTGTTACACAAGTGTGCCATTGCCCGCTGGCGAAGACTCGTGAGGTCCCATGCCGCGTGGAGTCGTCGTGGTCGTGCGGCGGCGCGTGCGGGCGCGTGCTGTCGTGCGGCGCGCACGTGTGCCGGAGCACGTGCCACGAGCCCCCCTGCCCCCCCTGCGACCTGCTGCCTGCCGCGCTCATCACATGTCCGTGCGGGAAGACTAGGTTAGTGTTATAA
Protein
MCSSKLPQVCGRPCQRMLQCGVHKCMKECHEGPCEDCTESVTQVCHCPLAKTREVPCRVESSWSCGGACGRVLSCGAHVCRSTCHEPPCPPCDLLPAALITCPCGKTRLVL
Summary
Similarity
Belongs to the NFX1 family.
Keywords
Alternative splicing
Complete proteome
DNA-binding
Metal-binding
Nucleus
Phosphoprotein
Reference proteome
Repeat
RNA-binding
Transcription
Transcription regulation
Zinc
Zinc-finger
Feature
chain Protein shuttle craft
splice variant In isoform B.
splice variant In isoform B.
Uniprot
H9J7Z2
A0A2A4K355
A0A2A4K2T6
A0A194QGR9
A0A194QXJ0
L7X1P9
+ More
A0A3S2LJM5 A0A2H1V9F9 A0A212EP74 D0AB98 E9AVJ1 A0A3B3HQ34 A0A3P9KF86 A0A3B3HBV7 A0A3P9KFR6 E9BFL4 A0A3P9LYK9 A0A3B3H9U3 E9AGY6 A0A3S5H7A4 T1IRV0 A0A139WHD1 D6WMN1 Q4QBY8 A0A2J7QGA5 A0A2J7QGB1 E9H0P6 A0A3R7MD54 E8NH79 P40798-2 A0A061IS94 M9PDG3 M9PD57 P40798 B4MZE5 V9IHK4 A0A3P9J2S4 A0A2V2US01 Q4D928 A0A026WYJ9 A0A1X0NUI9 V5BFS7 A0A2V2UNA9 A0A2V2WLN1 Q4E1D9 K2LTG1 A0A182MHE8 V5GY42 B3MM96 A0A0P5KGC1 A0A0P5VH86 A0A0N8E1S9 A0A0P5KFL6 A0A3P3Z6Q5 A4HC58 A0A0P5U8W2 A0A088S9I8 A0A0P5PEI5 A0A3B3CWX9 B4NYD7 Q29P77 B4GK89 A0A1Y1JZ76 A0A0P5YM02 A0A1Y2CMK2 A0A3B3B700 B4KLB0 A0A0P5LKS5 A0A0K9PAU4 A0A0L0CJR7 A0A0P5B1I7 A0A3L6PGY4 A0A0P5QU35 A0A0N0VFZ7 A0A1W4WC55 A0A0N4UW62 A0A0P4ZYX1 A0A2R5LJK7 A0A0P5S2J5 A0A0P5FMM2 A0A0P5XCR0 A0A0P5XSZ8 B4HXM6 A0A0P5Q5B7 D8LYV5 B3N5P4 B4Q602 A0A2P8Z3T6 A0A0L7RDI2 A0A0N8DCJ5 A0A0P5AUP1 A0A0P5A3Q8 A0A0N8EQ75 A0A0P5SWI5 K3XV33 A0A0J9R2N5 A0A162CCJ7 A0A0P5A851 A0A0P5DJZ2
A0A3S2LJM5 A0A2H1V9F9 A0A212EP74 D0AB98 E9AVJ1 A0A3B3HQ34 A0A3P9KF86 A0A3B3HBV7 A0A3P9KFR6 E9BFL4 A0A3P9LYK9 A0A3B3H9U3 E9AGY6 A0A3S5H7A4 T1IRV0 A0A139WHD1 D6WMN1 Q4QBY8 A0A2J7QGA5 A0A2J7QGB1 E9H0P6 A0A3R7MD54 E8NH79 P40798-2 A0A061IS94 M9PDG3 M9PD57 P40798 B4MZE5 V9IHK4 A0A3P9J2S4 A0A2V2US01 Q4D928 A0A026WYJ9 A0A1X0NUI9 V5BFS7 A0A2V2UNA9 A0A2V2WLN1 Q4E1D9 K2LTG1 A0A182MHE8 V5GY42 B3MM96 A0A0P5KGC1 A0A0P5VH86 A0A0N8E1S9 A0A0P5KFL6 A0A3P3Z6Q5 A4HC58 A0A0P5U8W2 A0A088S9I8 A0A0P5PEI5 A0A3B3CWX9 B4NYD7 Q29P77 B4GK89 A0A1Y1JZ76 A0A0P5YM02 A0A1Y2CMK2 A0A3B3B700 B4KLB0 A0A0P5LKS5 A0A0K9PAU4 A0A0L0CJR7 A0A0P5B1I7 A0A3L6PGY4 A0A0P5QU35 A0A0N0VFZ7 A0A1W4WC55 A0A0N4UW62 A0A0P4ZYX1 A0A2R5LJK7 A0A0P5S2J5 A0A0P5FMM2 A0A0P5XCR0 A0A0P5XSZ8 B4HXM6 A0A0P5Q5B7 D8LYV5 B3N5P4 B4Q602 A0A2P8Z3T6 A0A0L7RDI2 A0A0N8DCJ5 A0A0P5AUP1 A0A0P5A3Q8 A0A0N8EQ75 A0A0P5SWI5 K3XV33 A0A0J9R2N5 A0A162CCJ7 A0A0P5A851 A0A0P5DJZ2
Pubmed
19121390
26354079
23674305
22118469
22038252
17554307
+ More
22038251 17572675 30409989 18362917 19820115 16020728 21292972 30355302 8524296 10471707 10731132 12537572 10731138 18327897 12537568 12537573 12537574 16110336 17569856 17569867 17994087 29708484 16020725 24508170 24482508 23035642 25707621 29451363 17550304 15632085 28004739 26814964 26108605 30683860 29403074 22580951 22936249
22038251 17572675 30409989 18362917 19820115 16020728 21292972 30355302 8524296 10471707 10731132 12537572 10731138 18327897 12537568 12537573 12537574 16110336 17569856 17569867 17994087 29708484 16020725 24508170 24482508 23035642 25707621 29451363 17550304 15632085 28004739 26814964 26108605 30683860 29403074 22580951 22936249
EMBL
BABH01020191
BABH01020192
NWSH01000227
PCG78213.1
PCG78214.1
KQ459232
+ More
KPJ02641.1 KQ461155 KPJ08291.1 KC469893 AGC92703.2 RSAL01000090 RVE48091.1 ODYU01001380 SOQ37468.1 AGBW02013517 OWR43279.1 FP102340 CBH09293.1 FR799575 CBZ26973.1 FR799609 CBZ34040.1 FR796454 UINB01000022 CBZ08650.1 SUZ41696.1 CP029521 AYU78699.1 JH431388 KQ971343 KYB27326.1 EFA04758.2 FR796418 CAJ03782.1 NEVH01014379 PNF27625.1 PNF27624.1 GL732581 EFX74683.1 MKGL01000327 RNF00413.1 BT125955 ADX35934.1 U09306 AE014134 AF145679 AUPL01006715 ESL05628.1 AGB92995.1 AGB92996.1 CH963913 EDW77484.2 JR046058 AEY60122.1 PRFC01000577 PWU85996.1 AAHK01000795 EAN89027.1 KK107064 EZA60846.1 NBCO01000019 ORC87859.1 AYLP01000076 ESS64932.1 PRFA01000142 MKKV01000219 PWU85837.1 RNF12979.1 PRFC01000083 MKQG01002482 PWV09102.1 RNC44062.1 AAHK01000050 EAN98570.1 AHKC01022676 EKF26023.1 AXCM01000660 GALX01003148 JAB65318.1 CH902620 EDV31856.1 GDIQ01210183 JAK41542.1 GDIP01115026 JAL88688.1 GDIQ01070236 JAN24501.1 GDIQ01191536 JAK60189.1 LS997621 SYZ65832.1 FR798997 CAM45049.2 GDIP01116421 JAL87293.1 CP009391 AIN98301.1 GDIQ01135765 JAL15961.1 CM000157 EDW89773.1 CH379058 EAL34416.2 CH479184 EDW37055.1 GEZM01096724 JAV54622.1 GDIP01055985 JAM47730.1 MCGO01000012 ORY48177.1 CH933807 EDW11771.1 GDIQ01172193 JAK79532.1 LFYR01001054 KMZ65305.1 JRES01000305 KNC32491.1 GDIP01191112 JAJ32290.1 PQIB02000018 RLM56259.1 GDIQ01119112 JAL32614.1 LGTL01000006 KPA81944.1 UXUI01007208 VDD86277.1 GDIP01206262 JAJ17140.1 GGLE01005607 MBY09733.1 GDIQ01102271 JAL49455.1 GDIQ01254719 JAJ97005.1 GDIP01074037 JAM29678.1 GDIP01067825 JAM35890.1 CH480818 EDW51806.1 GDIQ01128831 JAL22895.1 FN668640 CBK20994.2 CH954177 EDV58003.1 CM000361 EDX05105.1 PYGN01000208 PSN51156.1 KQ414614 KOC68840.1 GDIP01047087 JAM56628.1 GDIP01208014 JAJ15388.1 GDIP01207964 JAJ15438.1 GDIQ01004668 JAN90069.1 GDIP01139280 JAL64434.1 AGNK02002331 CM003531 RCV21105.1 CM002910 KMY90343.1 LRGB01001005 KZS14152.1 GDIP01206263 JAJ17139.1 GDIP01158477 JAJ64925.1
KPJ02641.1 KQ461155 KPJ08291.1 KC469893 AGC92703.2 RSAL01000090 RVE48091.1 ODYU01001380 SOQ37468.1 AGBW02013517 OWR43279.1 FP102340 CBH09293.1 FR799575 CBZ26973.1 FR799609 CBZ34040.1 FR796454 UINB01000022 CBZ08650.1 SUZ41696.1 CP029521 AYU78699.1 JH431388 KQ971343 KYB27326.1 EFA04758.2 FR796418 CAJ03782.1 NEVH01014379 PNF27625.1 PNF27624.1 GL732581 EFX74683.1 MKGL01000327 RNF00413.1 BT125955 ADX35934.1 U09306 AE014134 AF145679 AUPL01006715 ESL05628.1 AGB92995.1 AGB92996.1 CH963913 EDW77484.2 JR046058 AEY60122.1 PRFC01000577 PWU85996.1 AAHK01000795 EAN89027.1 KK107064 EZA60846.1 NBCO01000019 ORC87859.1 AYLP01000076 ESS64932.1 PRFA01000142 MKKV01000219 PWU85837.1 RNF12979.1 PRFC01000083 MKQG01002482 PWV09102.1 RNC44062.1 AAHK01000050 EAN98570.1 AHKC01022676 EKF26023.1 AXCM01000660 GALX01003148 JAB65318.1 CH902620 EDV31856.1 GDIQ01210183 JAK41542.1 GDIP01115026 JAL88688.1 GDIQ01070236 JAN24501.1 GDIQ01191536 JAK60189.1 LS997621 SYZ65832.1 FR798997 CAM45049.2 GDIP01116421 JAL87293.1 CP009391 AIN98301.1 GDIQ01135765 JAL15961.1 CM000157 EDW89773.1 CH379058 EAL34416.2 CH479184 EDW37055.1 GEZM01096724 JAV54622.1 GDIP01055985 JAM47730.1 MCGO01000012 ORY48177.1 CH933807 EDW11771.1 GDIQ01172193 JAK79532.1 LFYR01001054 KMZ65305.1 JRES01000305 KNC32491.1 GDIP01191112 JAJ32290.1 PQIB02000018 RLM56259.1 GDIQ01119112 JAL32614.1 LGTL01000006 KPA81944.1 UXUI01007208 VDD86277.1 GDIP01206262 JAJ17140.1 GGLE01005607 MBY09733.1 GDIQ01102271 JAL49455.1 GDIQ01254719 JAJ97005.1 GDIP01074037 JAM29678.1 GDIP01067825 JAM35890.1 CH480818 EDW51806.1 GDIQ01128831 JAL22895.1 FN668640 CBK20994.2 CH954177 EDV58003.1 CM000361 EDX05105.1 PYGN01000208 PSN51156.1 KQ414614 KOC68840.1 GDIP01047087 JAM56628.1 GDIP01208014 JAJ15388.1 GDIP01207964 JAJ15438.1 GDIQ01004668 JAN90069.1 GDIP01139280 JAL64434.1 AGNK02002331 CM003531 RCV21105.1 CM002910 KMY90343.1 LRGB01001005 KZS14152.1 GDIP01206263 JAJ17139.1 GDIP01158477 JAJ64925.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000283053
UP000007151
+ More
UP000007259 UP000001038 UP000265180 UP000008980 UP000008153 UP000274082 UP000007266 UP000000542 UP000235965 UP000000305 UP000000803 UP000007798 UP000265200 UP000002296 UP000053097 UP000192257 UP000075883 UP000007801 UP000007258 UP000261560 UP000002282 UP000001819 UP000008744 UP000193642 UP000009192 UP000036987 UP000037069 UP000275267 UP000192221 UP000038041 UP000274131 UP000001292 UP000008711 UP000000304 UP000245037 UP000053825 UP000004995 UP000076858
UP000007259 UP000001038 UP000265180 UP000008980 UP000008153 UP000274082 UP000007266 UP000000542 UP000235965 UP000000305 UP000000803 UP000007798 UP000265200 UP000002296 UP000053097 UP000192257 UP000075883 UP000007801 UP000007258 UP000261560 UP000002282 UP000001819 UP000008744 UP000193642 UP000009192 UP000036987 UP000037069 UP000275267 UP000192221 UP000038041 UP000274131 UP000001292 UP000008711 UP000000304 UP000245037 UP000053825 UP000004995 UP000076858
Interpro
Gene 3D
ProteinModelPortal
H9J7Z2
A0A2A4K355
A0A2A4K2T6
A0A194QGR9
A0A194QXJ0
L7X1P9
+ More
A0A3S2LJM5 A0A2H1V9F9 A0A212EP74 D0AB98 E9AVJ1 A0A3B3HQ34 A0A3P9KF86 A0A3B3HBV7 A0A3P9KFR6 E9BFL4 A0A3P9LYK9 A0A3B3H9U3 E9AGY6 A0A3S5H7A4 T1IRV0 A0A139WHD1 D6WMN1 Q4QBY8 A0A2J7QGA5 A0A2J7QGB1 E9H0P6 A0A3R7MD54 E8NH79 P40798-2 A0A061IS94 M9PDG3 M9PD57 P40798 B4MZE5 V9IHK4 A0A3P9J2S4 A0A2V2US01 Q4D928 A0A026WYJ9 A0A1X0NUI9 V5BFS7 A0A2V2UNA9 A0A2V2WLN1 Q4E1D9 K2LTG1 A0A182MHE8 V5GY42 B3MM96 A0A0P5KGC1 A0A0P5VH86 A0A0N8E1S9 A0A0P5KFL6 A0A3P3Z6Q5 A4HC58 A0A0P5U8W2 A0A088S9I8 A0A0P5PEI5 A0A3B3CWX9 B4NYD7 Q29P77 B4GK89 A0A1Y1JZ76 A0A0P5YM02 A0A1Y2CMK2 A0A3B3B700 B4KLB0 A0A0P5LKS5 A0A0K9PAU4 A0A0L0CJR7 A0A0P5B1I7 A0A3L6PGY4 A0A0P5QU35 A0A0N0VFZ7 A0A1W4WC55 A0A0N4UW62 A0A0P4ZYX1 A0A2R5LJK7 A0A0P5S2J5 A0A0P5FMM2 A0A0P5XCR0 A0A0P5XSZ8 B4HXM6 A0A0P5Q5B7 D8LYV5 B3N5P4 B4Q602 A0A2P8Z3T6 A0A0L7RDI2 A0A0N8DCJ5 A0A0P5AUP1 A0A0P5A3Q8 A0A0N8EQ75 A0A0P5SWI5 K3XV33 A0A0J9R2N5 A0A162CCJ7 A0A0P5A851 A0A0P5DJZ2
A0A3S2LJM5 A0A2H1V9F9 A0A212EP74 D0AB98 E9AVJ1 A0A3B3HQ34 A0A3P9KF86 A0A3B3HBV7 A0A3P9KFR6 E9BFL4 A0A3P9LYK9 A0A3B3H9U3 E9AGY6 A0A3S5H7A4 T1IRV0 A0A139WHD1 D6WMN1 Q4QBY8 A0A2J7QGA5 A0A2J7QGB1 E9H0P6 A0A3R7MD54 E8NH79 P40798-2 A0A061IS94 M9PDG3 M9PD57 P40798 B4MZE5 V9IHK4 A0A3P9J2S4 A0A2V2US01 Q4D928 A0A026WYJ9 A0A1X0NUI9 V5BFS7 A0A2V2UNA9 A0A2V2WLN1 Q4E1D9 K2LTG1 A0A182MHE8 V5GY42 B3MM96 A0A0P5KGC1 A0A0P5VH86 A0A0N8E1S9 A0A0P5KFL6 A0A3P3Z6Q5 A4HC58 A0A0P5U8W2 A0A088S9I8 A0A0P5PEI5 A0A3B3CWX9 B4NYD7 Q29P77 B4GK89 A0A1Y1JZ76 A0A0P5YM02 A0A1Y2CMK2 A0A3B3B700 B4KLB0 A0A0P5LKS5 A0A0K9PAU4 A0A0L0CJR7 A0A0P5B1I7 A0A3L6PGY4 A0A0P5QU35 A0A0N0VFZ7 A0A1W4WC55 A0A0N4UW62 A0A0P4ZYX1 A0A2R5LJK7 A0A0P5S2J5 A0A0P5FMM2 A0A0P5XCR0 A0A0P5XSZ8 B4HXM6 A0A0P5Q5B7 D8LYV5 B3N5P4 B4Q602 A0A2P8Z3T6 A0A0L7RDI2 A0A0N8DCJ5 A0A0P5AUP1 A0A0P5A3Q8 A0A0N8EQ75 A0A0P5SWI5 K3XV33 A0A0J9R2N5 A0A162CCJ7 A0A0P5A851 A0A0P5DJZ2
Ontologies
GO
PANTHER
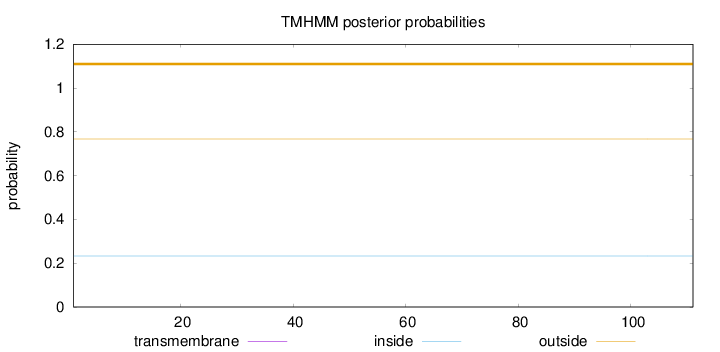
Topology
Subcellular location
Nucleus
Length:
111
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.23314
outside
1 - 111
Population Genetic Test Statistics
Pi
133.108066
Theta
122.960012
Tajima's D
0.366195
CLR
0
CSRT
0.474076296185191
Interpretation
Uncertain
