Gene
KWMTBOMO09500
Annotation
PREDICTED:_ceramide_synthase_6-like_isoform_X1_[Bombyx_mori]
Transcription factor
Location in the cell
PlasmaMembrane Reliability : 4.202
Sequence
CDS
ATGAATACGTTACGATTATTATTAGACACGTTTTGGGATGAATCTGTTTGGTTGCCTCCTAATACTACATGGGAAGATCTGGAGCCGGGTCCTGACAAAGTCGTGATTTACAATGACTACCGGCATCTTTTTTATCCGTTGCCGCTGGCGTTGGTGCTTATAGTATTACGGCATGTACTGGAGAAGTACTGGTTTGCACCGTTTGGAAAATCAATAGGTATTAAAAATACCAGACCAAAGAAAGCACCTAACAATCCTAAATTAGAAAATGCATATCAAGCATCGTCCAAACTGAAACATAAACAGATATGTGCACTGGCGAAGCAAGTGGACTTATCGGAGAGACAGGTGGAAAGATGGTGGCGTTTACGTCGCAGCCAAGACAAGCCCTCCACCCTGGTCAAGTTCTGCGAGAACTGTTGGAGATGCAGCTTCTATTTGTACAACTTCTCGTACGGTATGTTCATACTATGGGAGAAAACTTGGCTGTGGGATATCGACCAGTGCTACATTGGCTATCCCCATCAGGGCGTGACCGGTGACGTGTGGTGGTACTACATGATATCGTCCGCCTTCTATTGGTCGCTGACCATCTCGCAGTTCTGGGACGTCAAGCGCAAGGACTTCTGGCAGATGTTCGTCCACCACATCGCCACCATAGCGCTGCTCTCGTTCAGCTGGGTCTGCAACCTGCACCGGATCGGAACCCTGGTTCTGCTGACGCACGACTGCGCGGACATATTCTTGGAGGCGGCCAAGGCCACGAAATACGCGAGCTACCAGAAGCTATGCGATTTCCTTTTCACGATCTTCACTCTGTTATGGATAGTGACGAGGAATGGTATTTATCCGTTCTATATTATTTGGAGCACGTCAATCCGAGCACCGATGTTGCTTCCAATGTTCCCTGCCTATTACATATTCAATTCCCTGCTCTGTTTGCTGTTGGCCCTGCACATGGTTTGGACCTGGCTCATAGTTCAAGTCGCCTACAAGTCCATAAGCGCCGGGCAGATGGAAGGCGACATTAGAAGCTCCAGTTCGGATATCAGTGACAGCTCTCATCATTCAAACCACAGCACGCCCAATCGCGTTCGAAGTAAAAAAGACGCGTAA
Protein
MNTLRLLLDTFWDESVWLPPNTTWEDLEPGPDKVVIYNDYRHLFYPLPLALVLIVLRHVLEKYWFAPFGKSIGIKNTRPKKAPNNPKLENAYQASSKLKHKQICALAKQVDLSERQVERWWRLRRSQDKPSTLVKFCENCWRCSFYLYNFSYGMFILWEKTWLWDIDQCYIGYPHQGVTGDVWWYYMISSAFYWSLTISQFWDVKRKDFWQMFVHHIATIALLSFSWVCNLHRIGTLVLLTHDCADIFLEAAKATKYASYQKLCDFLFTIFTLLWIVTRNGIYPFYIIWSTSIRAPMLLPMFPAYYIFNSLLCLLLALHMVWTWLIVQVAYKSISAGQMEGDIRSSSSDISDSSHHSNHSTPNRVRSKKDA
Summary
Uniprot
A0A1E1VY27
A0A1E1W2Q5
A0A2A4JIT0
A0A1E1WFG3
A0A1E1W3C8
A0A3S2LL92
+ More
A0A2A4JGY2 A0A2W1BHT5 A0A437BFD0 A0A2H1VPH1 A0A194Q6Q3 A0A1L8DUL0 A0A336MAZ5 A0A1Q3G272 E2C5U9 A0A1Q3G2A4 A0A1Q3G277 A0A1Q3G2E4 D6WYR1 A0A1B0FGD7 A0A1B0BJV0 A0A1A9VC72 A0A0L7R4X8 A0A1I8MJC5 A0A067RQ48 A0A026WNX4 W8C680 A0A154PDG5 A0A182V3Z5 Q7PY62 A0A2C9GPW4 U5EU05 A0A2C9H886 Q16WU6 A0A158NR19 A0A182QKD2 E2ADT5 K7IN59 A0A195E5N7 A0A195B130 F4WP94 A0A182G1G4 B4L5F1 A0A0M3QYZ5 A0A1Y1K4Z9 A0A182PQZ9 B4JNE6 A0A2M4CIX5 A0A0A1WHQ7 A0A2M4CIV4 Q16WU5 T1DK73 A0A0K8U7V5 B4MAX9 A0A2R7VUW9 A0A2M4AM46 A0A2M4BQA1 A0A2M4BQ26 A0A2M4AMM7 A0A0C9REG6 A0A1W4VFJ0 A0A2M3Z6W5 A0A2M3Z6T8 A0A0P8YKM5 A0A0Q9WDH8 Q29GF6 A0A232F7B8 B4NE76 B4I0D3 B4HB90 Q9W423 B4PYQ4 A0A3B0KEC9 B3NXQ1 A0A2A3E101 R4G7Z2 A0A151ICM1 A0A195EUJ7 A0A2S2P1G1 A0A1L8EBK0 C4WXB7 A0A1L8EB48 A0A1B6DSL3 A0A023F8K1 A0A0V0GAE7 A0A224XSH7 A0A069DTC4 J9K7R9 A0A1B6DU08 A0A0A9XX01 V5GK93 A0A146L0Z9 A0A1B6KN89 A0A1B6F8B2 A0A1B6IBE6 T1E2R0 A0A2W1BKB4 A0A1B0CIB7 A0A0N1IFL4 A0A2H1VPF7
A0A2A4JGY2 A0A2W1BHT5 A0A437BFD0 A0A2H1VPH1 A0A194Q6Q3 A0A1L8DUL0 A0A336MAZ5 A0A1Q3G272 E2C5U9 A0A1Q3G2A4 A0A1Q3G277 A0A1Q3G2E4 D6WYR1 A0A1B0FGD7 A0A1B0BJV0 A0A1A9VC72 A0A0L7R4X8 A0A1I8MJC5 A0A067RQ48 A0A026WNX4 W8C680 A0A154PDG5 A0A182V3Z5 Q7PY62 A0A2C9GPW4 U5EU05 A0A2C9H886 Q16WU6 A0A158NR19 A0A182QKD2 E2ADT5 K7IN59 A0A195E5N7 A0A195B130 F4WP94 A0A182G1G4 B4L5F1 A0A0M3QYZ5 A0A1Y1K4Z9 A0A182PQZ9 B4JNE6 A0A2M4CIX5 A0A0A1WHQ7 A0A2M4CIV4 Q16WU5 T1DK73 A0A0K8U7V5 B4MAX9 A0A2R7VUW9 A0A2M4AM46 A0A2M4BQA1 A0A2M4BQ26 A0A2M4AMM7 A0A0C9REG6 A0A1W4VFJ0 A0A2M3Z6W5 A0A2M3Z6T8 A0A0P8YKM5 A0A0Q9WDH8 Q29GF6 A0A232F7B8 B4NE76 B4I0D3 B4HB90 Q9W423 B4PYQ4 A0A3B0KEC9 B3NXQ1 A0A2A3E101 R4G7Z2 A0A151ICM1 A0A195EUJ7 A0A2S2P1G1 A0A1L8EBK0 C4WXB7 A0A1L8EB48 A0A1B6DSL3 A0A023F8K1 A0A0V0GAE7 A0A224XSH7 A0A069DTC4 J9K7R9 A0A1B6DU08 A0A0A9XX01 V5GK93 A0A146L0Z9 A0A1B6KN89 A0A1B6F8B2 A0A1B6IBE6 T1E2R0 A0A2W1BKB4 A0A1B0CIB7 A0A0N1IFL4 A0A2H1VPF7
Pubmed
28756777
26354079
20798317
18362917
19820115
25315136
+ More
24845553 24508170 24495485 12364791 14747013 17210077 17510324 21347285 20075255 21719571 26483478 17994087 28004739 25830018 18057021 15632085 28648823 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25474469 26334808 25401762 26823975 24330624
24845553 24508170 24495485 12364791 14747013 17210077 17510324 21347285 20075255 21719571 26483478 17994087 28004739 25830018 18057021 15632085 28648823 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25474469 26334808 25401762 26823975 24330624
EMBL
GDQN01011470
JAT79584.1
GDQN01009798
GDQN01001145
JAT81256.1
JAT89909.1
+ More
NWSH01001411 PCG71333.1 GDQN01005366 JAT85688.1 GDQN01009574 JAT81480.1 RSAL01000071 RVE49134.1 PCG71335.1 KZ150116 PZC73275.1 RVE49133.1 ODYU01003668 SOQ42723.1 KQ459582 KPI99070.1 GFDF01003974 JAV10110.1 UFQS01000320 UFQT01000320 SSX02850.1 SSX23218.1 GFDL01001117 JAV33928.1 GL452834 EFN76685.1 GFDL01001113 JAV33932.1 GFDL01001142 JAV33903.1 GFDL01001119 JAV33926.1 KQ971357 EFA08405.2 CCAG010008765 JXJN01015636 JXJN01015637 KQ414657 KOC65826.1 KK852531 KDR21874.1 KK107139 EZA57715.1 GAMC01001277 JAC05279.1 KQ434879 KZC09949.1 AAAB01008987 EAA01235.5 APCN01000913 GANO01002545 JAB57326.1 CH477554 EAT39065.1 ADTU01023745 ADTU01023746 AXCN02000314 GL438820 EFN68447.1 KQ979608 KYN20478.1 KQ976691 KYM77909.1 GL888243 EGI64075.1 JXUM01038484 JXUM01038485 JXUM01038486 KQ561167 KXJ79489.1 CH933811 EDW06410.1 CP012528 ALC48479.1 GEZM01092745 JAV56473.1 CH916371 EDV92239.1 GGFL01001118 MBW65296.1 GBXI01015910 JAC98381.1 GGFL01001114 MBW65292.1 EAT39066.1 GAMD01001086 JAB00505.1 GDHF01029595 GDHF01024706 JAI22719.1 JAI27608.1 CH940655 EDW66388.1 KK854101 PTY11296.1 GGFK01008520 MBW41841.1 GGFJ01006043 MBW55184.1 GGFJ01006044 MBW55185.1 GGFK01008537 MBW41858.1 GBYB01011557 JAG81324.1 GGFM01003479 MBW24230.1 GGFM01003486 MBW24237.1 CH902621 KPU81618.1 KRF82662.1 KRF82663.1 KRF82664.1 CH379064 EAL32153.2 NNAY01000810 OXU26390.1 CH964239 EDW82045.1 CH480819 EDW52964.1 CH479248 EDW37902.1 AE014298 AY061255 AY066020 AAF46137.2 AAG22409.2 AAL28803.1 AAL57756.1 CM000162 EDX00990.1 OUUW01000011 SPP86670.1 CH954180 EDV47352.1 KZ288468 PBC25378.1 ACPB03006224 GAHY01001877 JAA75633.1 KQ978052 KYM97789.1 KQ981965 KYN31831.1 GGMR01010684 MBY23303.1 GFDG01002810 JAV15989.1 AK342547 BAH72537.1 GFDG01002809 JAV15990.1 GEDC01008653 JAS28645.1 GBBI01001001 JAC17711.1 GECL01000985 JAP05139.1 GFTR01005495 JAW10931.1 GBGD01001888 JAC87001.1 ABLF02015011 GEDC01008131 JAS29167.1 GBHO01021809 GBRD01010463 GBRD01010462 GDHC01003308 JAG21795.1 JAG55361.1 JAQ15321.1 GALX01003992 JAB64474.1 GDHC01016990 JAQ01639.1 GEBQ01027049 JAT12928.1 GECZ01023343 JAS46426.1 GECU01023441 JAS84265.1 GALA01001070 JAA93782.1 PZC73276.1 AJWK01013182 AJWK01013183 AJWK01013184 KQ460651 KPJ13010.1 SOQ42721.1
NWSH01001411 PCG71333.1 GDQN01005366 JAT85688.1 GDQN01009574 JAT81480.1 RSAL01000071 RVE49134.1 PCG71335.1 KZ150116 PZC73275.1 RVE49133.1 ODYU01003668 SOQ42723.1 KQ459582 KPI99070.1 GFDF01003974 JAV10110.1 UFQS01000320 UFQT01000320 SSX02850.1 SSX23218.1 GFDL01001117 JAV33928.1 GL452834 EFN76685.1 GFDL01001113 JAV33932.1 GFDL01001142 JAV33903.1 GFDL01001119 JAV33926.1 KQ971357 EFA08405.2 CCAG010008765 JXJN01015636 JXJN01015637 KQ414657 KOC65826.1 KK852531 KDR21874.1 KK107139 EZA57715.1 GAMC01001277 JAC05279.1 KQ434879 KZC09949.1 AAAB01008987 EAA01235.5 APCN01000913 GANO01002545 JAB57326.1 CH477554 EAT39065.1 ADTU01023745 ADTU01023746 AXCN02000314 GL438820 EFN68447.1 KQ979608 KYN20478.1 KQ976691 KYM77909.1 GL888243 EGI64075.1 JXUM01038484 JXUM01038485 JXUM01038486 KQ561167 KXJ79489.1 CH933811 EDW06410.1 CP012528 ALC48479.1 GEZM01092745 JAV56473.1 CH916371 EDV92239.1 GGFL01001118 MBW65296.1 GBXI01015910 JAC98381.1 GGFL01001114 MBW65292.1 EAT39066.1 GAMD01001086 JAB00505.1 GDHF01029595 GDHF01024706 JAI22719.1 JAI27608.1 CH940655 EDW66388.1 KK854101 PTY11296.1 GGFK01008520 MBW41841.1 GGFJ01006043 MBW55184.1 GGFJ01006044 MBW55185.1 GGFK01008537 MBW41858.1 GBYB01011557 JAG81324.1 GGFM01003479 MBW24230.1 GGFM01003486 MBW24237.1 CH902621 KPU81618.1 KRF82662.1 KRF82663.1 KRF82664.1 CH379064 EAL32153.2 NNAY01000810 OXU26390.1 CH964239 EDW82045.1 CH480819 EDW52964.1 CH479248 EDW37902.1 AE014298 AY061255 AY066020 AAF46137.2 AAG22409.2 AAL28803.1 AAL57756.1 CM000162 EDX00990.1 OUUW01000011 SPP86670.1 CH954180 EDV47352.1 KZ288468 PBC25378.1 ACPB03006224 GAHY01001877 JAA75633.1 KQ978052 KYM97789.1 KQ981965 KYN31831.1 GGMR01010684 MBY23303.1 GFDG01002810 JAV15989.1 AK342547 BAH72537.1 GFDG01002809 JAV15990.1 GEDC01008653 JAS28645.1 GBBI01001001 JAC17711.1 GECL01000985 JAP05139.1 GFTR01005495 JAW10931.1 GBGD01001888 JAC87001.1 ABLF02015011 GEDC01008131 JAS29167.1 GBHO01021809 GBRD01010463 GBRD01010462 GDHC01003308 JAG21795.1 JAG55361.1 JAQ15321.1 GALX01003992 JAB64474.1 GDHC01016990 JAQ01639.1 GEBQ01027049 JAT12928.1 GECZ01023343 JAS46426.1 GECU01023441 JAS84265.1 GALA01001070 JAA93782.1 PZC73276.1 AJWK01013182 AJWK01013183 AJWK01013184 KQ460651 KPJ13010.1 SOQ42721.1
Proteomes
UP000218220
UP000283053
UP000053268
UP000008237
UP000007266
UP000092444
+ More
UP000092460 UP000078200 UP000053825 UP000095301 UP000027135 UP000053097 UP000076502 UP000075903 UP000007062 UP000075840 UP000076407 UP000008820 UP000005205 UP000075886 UP000000311 UP000002358 UP000078492 UP000078540 UP000007755 UP000069940 UP000249989 UP000009192 UP000092553 UP000075885 UP000001070 UP000008792 UP000192221 UP000007801 UP000001819 UP000215335 UP000007798 UP000001292 UP000008744 UP000000803 UP000002282 UP000268350 UP000008711 UP000242457 UP000015103 UP000078542 UP000078541 UP000007819 UP000092461 UP000053240
UP000092460 UP000078200 UP000053825 UP000095301 UP000027135 UP000053097 UP000076502 UP000075903 UP000007062 UP000075840 UP000076407 UP000008820 UP000005205 UP000075886 UP000000311 UP000002358 UP000078492 UP000078540 UP000007755 UP000069940 UP000249989 UP000009192 UP000092553 UP000075885 UP000001070 UP000008792 UP000192221 UP000007801 UP000001819 UP000215335 UP000007798 UP000001292 UP000008744 UP000000803 UP000002282 UP000268350 UP000008711 UP000242457 UP000015103 UP000078542 UP000078541 UP000007819 UP000092461 UP000053240
Interpro
SUPFAM
SSF46689
SSF46689
ProteinModelPortal
A0A1E1VY27
A0A1E1W2Q5
A0A2A4JIT0
A0A1E1WFG3
A0A1E1W3C8
A0A3S2LL92
+ More
A0A2A4JGY2 A0A2W1BHT5 A0A437BFD0 A0A2H1VPH1 A0A194Q6Q3 A0A1L8DUL0 A0A336MAZ5 A0A1Q3G272 E2C5U9 A0A1Q3G2A4 A0A1Q3G277 A0A1Q3G2E4 D6WYR1 A0A1B0FGD7 A0A1B0BJV0 A0A1A9VC72 A0A0L7R4X8 A0A1I8MJC5 A0A067RQ48 A0A026WNX4 W8C680 A0A154PDG5 A0A182V3Z5 Q7PY62 A0A2C9GPW4 U5EU05 A0A2C9H886 Q16WU6 A0A158NR19 A0A182QKD2 E2ADT5 K7IN59 A0A195E5N7 A0A195B130 F4WP94 A0A182G1G4 B4L5F1 A0A0M3QYZ5 A0A1Y1K4Z9 A0A182PQZ9 B4JNE6 A0A2M4CIX5 A0A0A1WHQ7 A0A2M4CIV4 Q16WU5 T1DK73 A0A0K8U7V5 B4MAX9 A0A2R7VUW9 A0A2M4AM46 A0A2M4BQA1 A0A2M4BQ26 A0A2M4AMM7 A0A0C9REG6 A0A1W4VFJ0 A0A2M3Z6W5 A0A2M3Z6T8 A0A0P8YKM5 A0A0Q9WDH8 Q29GF6 A0A232F7B8 B4NE76 B4I0D3 B4HB90 Q9W423 B4PYQ4 A0A3B0KEC9 B3NXQ1 A0A2A3E101 R4G7Z2 A0A151ICM1 A0A195EUJ7 A0A2S2P1G1 A0A1L8EBK0 C4WXB7 A0A1L8EB48 A0A1B6DSL3 A0A023F8K1 A0A0V0GAE7 A0A224XSH7 A0A069DTC4 J9K7R9 A0A1B6DU08 A0A0A9XX01 V5GK93 A0A146L0Z9 A0A1B6KN89 A0A1B6F8B2 A0A1B6IBE6 T1E2R0 A0A2W1BKB4 A0A1B0CIB7 A0A0N1IFL4 A0A2H1VPF7
A0A2A4JGY2 A0A2W1BHT5 A0A437BFD0 A0A2H1VPH1 A0A194Q6Q3 A0A1L8DUL0 A0A336MAZ5 A0A1Q3G272 E2C5U9 A0A1Q3G2A4 A0A1Q3G277 A0A1Q3G2E4 D6WYR1 A0A1B0FGD7 A0A1B0BJV0 A0A1A9VC72 A0A0L7R4X8 A0A1I8MJC5 A0A067RQ48 A0A026WNX4 W8C680 A0A154PDG5 A0A182V3Z5 Q7PY62 A0A2C9GPW4 U5EU05 A0A2C9H886 Q16WU6 A0A158NR19 A0A182QKD2 E2ADT5 K7IN59 A0A195E5N7 A0A195B130 F4WP94 A0A182G1G4 B4L5F1 A0A0M3QYZ5 A0A1Y1K4Z9 A0A182PQZ9 B4JNE6 A0A2M4CIX5 A0A0A1WHQ7 A0A2M4CIV4 Q16WU5 T1DK73 A0A0K8U7V5 B4MAX9 A0A2R7VUW9 A0A2M4AM46 A0A2M4BQA1 A0A2M4BQ26 A0A2M4AMM7 A0A0C9REG6 A0A1W4VFJ0 A0A2M3Z6W5 A0A2M3Z6T8 A0A0P8YKM5 A0A0Q9WDH8 Q29GF6 A0A232F7B8 B4NE76 B4I0D3 B4HB90 Q9W423 B4PYQ4 A0A3B0KEC9 B3NXQ1 A0A2A3E101 R4G7Z2 A0A151ICM1 A0A195EUJ7 A0A2S2P1G1 A0A1L8EBK0 C4WXB7 A0A1L8EB48 A0A1B6DSL3 A0A023F8K1 A0A0V0GAE7 A0A224XSH7 A0A069DTC4 J9K7R9 A0A1B6DU08 A0A0A9XX01 V5GK93 A0A146L0Z9 A0A1B6KN89 A0A1B6F8B2 A0A1B6IBE6 T1E2R0 A0A2W1BKB4 A0A1B0CIB7 A0A0N1IFL4 A0A2H1VPF7
PDB
2CQX
E-value=0.000532911,
Score=102
Ontologies
PATHWAY
GO
PANTHER
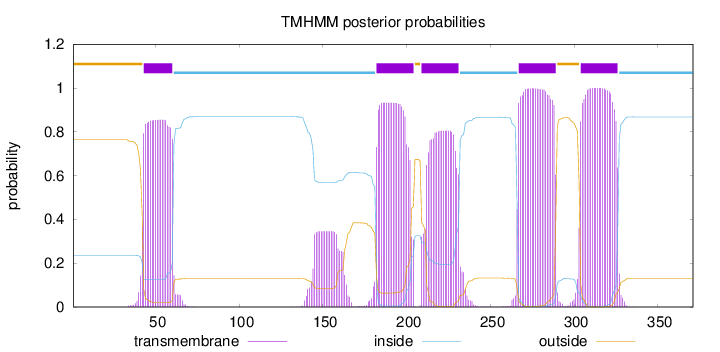
Topology
Subcellular location
Nucleus
Length:
371
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
105.16852
Exp number, first 60 AAs:
15.74489
Total prob of N-in:
0.23572
POSSIBLE N-term signal
sequence
outside
1 - 42
TMhelix
43 - 60
inside
61 - 181
TMhelix
182 - 204
outside
205 - 208
TMhelix
209 - 231
inside
232 - 266
TMhelix
267 - 289
outside
290 - 303
TMhelix
304 - 326
inside
327 - 371
Population Genetic Test Statistics
Pi
176.819795
Theta
163.921972
Tajima's D
0.014222
CLR
136.723506
CSRT
0.372281385930703
Interpretation
Uncertain
