Pre Gene Modal
BGIBMGA012776
Annotation
PREDICTED:_protein_FAM57A_[Bombyx_mori]
Transcription factor
Location in the cell
PlasmaMembrane Reliability : 4.786
Sequence
CDS
ATGGATCCAACATTTACTAAGCCCCAAAACCTTCGGCCAGCTATGTGGCTGTTATGCATTAGCAACGTGGCCTCACTCCTGGCTGCTCTCAACCTCGTCATCTGGCTGGAACCGGACGAGAGGATATCAATACCGCGAGGAAGTGCTTTAACAGTCCTGGGTCTGGTTTATTTCACGAGCCTGTTCGAGATCCTGAACGTGTGGGCTCTGTACACGGAGGCCGGCAGTCGGATCAGGCACCAGTACGCGCTGAGCCACGGAGACGTTTTGGACATCAACAACAAGCTGGTGTCTGCCGTCCAGGCGGTGTATTGCGTGGCGACGGGCGTGGTGGTGTGCAGCCACTCGTGCCGCCGCGACTTCATGCGCGCCTCCCACTACATGTCAGAGGCGTACGCGTGGTTCGGAGCCGCCTACTTCTTCTACGACATATGGTCCATGTACATGGTTCACATCCAGATGGTCTCCGGCGGGGAGTCGATGAAGGAGCAGGTCCGGAGGCGGGTGAGCGAGAGGAAGGCGCCGGTTTCGTTCCTCAAGTACTGCGCGCACGAGCCCGTCATACTGCTGCACCACGTCTTCATAGGGGGCTTCGGCTTCCTGGTCATTGTTCACCTGCGCGGCTCGCTGGGCGACTGCGCGTTCGGCTGGGTGTACGGCATGGAGGCGTCGACGCCGCTGGTGTGCGCGCGCGGCGTGCTGGCTCGCCTGCGCCTCAAGCGCTCGCCGCTCTACCTCGCCAACGGCCTGCTCATGCTGCTCACCTTCCTGCTGTGCCGCGTGCTGTCGCTGCCCTACGTGTGCTGGCTCTACAGCCGCGAGCTGGGACTCACCTACGCGCAGGCGATACAGTCGCTGCCGACGGGCTGCAAGATATCCATCTGCATCCTCCTGCTCCCGCAGCTGTACTGGTTCTACCTGATGTCCGCGGGGGCGCTGAAGATGCTGTTCCGCGCCGGCAAGCCCGCAGAGCTCGACGGCAAGGCGCCCCGGCCGACACCCCCGGCCGACAGGTCCGCCGGCGACGCGACCGAAGTGACCGACAAGCACAGTTGA
Protein
MDPTFTKPQNLRPAMWLLCISNVASLLAALNLVIWLEPDERISIPRGSALTVLGLVYFTSLFEILNVWALYTEAGSRIRHQYALSHGDVLDINNKLVSAVQAVYCVATGVVVCSHSCRRDFMRASHYMSEAYAWFGAAYFFYDIWSMYMVHIQMVSGGESMKEQVRRRVSERKAPVSFLKYCAHEPVILLHHVFIGGFGFLVIVHLRGSLGDCAFGWVYGMEASTPLVCARGVLARLRLKRSPLYLANGLLMLLTFLLCRVLSLPYVCWLYSRELGLTYAQAIQSLPTGCKISICILLLPQLYWFYLMSAGALKMLFRAGKPAELDGKAPRPTPPADRSAGDATEVTDKHS
Summary
Uniprot
H9JTB4
A0A2A4K2J8
A0A437BFA1
A0A194RSH8
A0A194Q0U5
D2A4Z1
+ More
V5I996 A0A0T6B029 A0A0L7LRY8 A0A1L8DK35 A0A1L8DJT1 A0A1L8DJW2 Q17PW3 A0A023ESG1 A0A1S4HDU9 A0A1Q3FE42 A0A182PI93 B0W0M1 A0A182HFX7 A0A182VB23 B4PXI3 A0A182Q1B9 Q9V3S7 A0A0K8US37 B3NW34 B4R7J4 A0A034W2K1 A0A182TNH3 A0A182YEC0 B3MY19 B4MJB1 A0A0A1WQL9 B4JKQ9 A0A2P8Y1G1 A0A182M8H0 A0A182SYF7 A0A182RCZ3 A0A0M4EIN8 A0A182WDE2 B4L2F1 A0A3B0K0C8 B4MEE5 A0A182JJ18 A0A336M8X9 A0A182MXI7 A0A1B0DA96 A0A1J1HP75 A0A182JSX4 E0VUB8 A0A182X5H9 Q29II1 A0A084W3I9 A0A1B0CSU3 B4H2I2 A0A1D2NG18 A0A226F756 A0A1S3CVZ4 A0A226EIS5 A0A1D2MZ95 A0A3Q3EPE1 A0A402EMB0 K1R9G4 A0A437C9X4
V5I996 A0A0T6B029 A0A0L7LRY8 A0A1L8DK35 A0A1L8DJT1 A0A1L8DJW2 Q17PW3 A0A023ESG1 A0A1S4HDU9 A0A1Q3FE42 A0A182PI93 B0W0M1 A0A182HFX7 A0A182VB23 B4PXI3 A0A182Q1B9 Q9V3S7 A0A0K8US37 B3NW34 B4R7J4 A0A034W2K1 A0A182TNH3 A0A182YEC0 B3MY19 B4MJB1 A0A0A1WQL9 B4JKQ9 A0A2P8Y1G1 A0A182M8H0 A0A182SYF7 A0A182RCZ3 A0A0M4EIN8 A0A182WDE2 B4L2F1 A0A3B0K0C8 B4MEE5 A0A182JJ18 A0A336M8X9 A0A182MXI7 A0A1B0DA96 A0A1J1HP75 A0A182JSX4 E0VUB8 A0A182X5H9 Q29II1 A0A084W3I9 A0A1B0CSU3 B4H2I2 A0A1D2NG18 A0A226F756 A0A1S3CVZ4 A0A226EIS5 A0A1D2MZ95 A0A3Q3EPE1 A0A402EMB0 K1R9G4 A0A437C9X4
Pubmed
EMBL
BABH01004172
NWSH01000261
PCG77910.1
RSAL01000071
RVE49103.1
KQ459700
+ More
KPJ20349.1 KQ459582 KPI98933.1 KQ971345 EFA05283.1 GALX01003320 JAB65146.1 LJIG01016367 KRT80815.1 JTDY01000224 KOB78207.1 GFDF01007374 JAV06710.1 GFDF01007372 JAV06712.1 GFDF01007373 JAV06711.1 CH477188 EAT48831.1 GAPW01001416 JAC12182.1 AAAB01008816 GFDL01009212 JAV25833.1 DS231818 EDS41323.1 APCN01005436 CM000162 EDX02934.1 AXCN02001714 AF181646 AE014298 AAD55432.1 AAF46579.1 GDHF01022800 JAI29514.1 CH954180 EDV46167.1 CM000366 EDX17553.1 GAKP01009161 JAC49791.1 CH902630 EDV38634.1 CH963738 EDW72200.2 GBXI01012973 JAD01319.1 CH916370 EDW00162.1 PYGN01001050 PSN38095.1 AXCM01008583 CP012528 ALC48804.1 CH933810 EDW06827.1 KRF93830.1 OUUW01000003 SPP78461.1 CH940664 EDW62920.1 KRF80855.1 UFQT01000258 SSX22498.1 AJVK01013239 CVRI01000014 CRK89755.1 DS235784 EEB16974.1 CH379063 EAL32672.2 ATLV01020054 KE525293 KFB44783.1 AJWK01026607 CH479204 EDW30549.1 LJIJ01000051 ODN04213.1 LNIX01000001 OXA65160.1 LNIX01000003 OXA57585.1 LJIJ01000363 ODM98332.1 BDOT01000024 GCF45299.1 JH817988 EKC30591.1 CM012455 RVE59572.1
KPJ20349.1 KQ459582 KPI98933.1 KQ971345 EFA05283.1 GALX01003320 JAB65146.1 LJIG01016367 KRT80815.1 JTDY01000224 KOB78207.1 GFDF01007374 JAV06710.1 GFDF01007372 JAV06712.1 GFDF01007373 JAV06711.1 CH477188 EAT48831.1 GAPW01001416 JAC12182.1 AAAB01008816 GFDL01009212 JAV25833.1 DS231818 EDS41323.1 APCN01005436 CM000162 EDX02934.1 AXCN02001714 AF181646 AE014298 AAD55432.1 AAF46579.1 GDHF01022800 JAI29514.1 CH954180 EDV46167.1 CM000366 EDX17553.1 GAKP01009161 JAC49791.1 CH902630 EDV38634.1 CH963738 EDW72200.2 GBXI01012973 JAD01319.1 CH916370 EDW00162.1 PYGN01001050 PSN38095.1 AXCM01008583 CP012528 ALC48804.1 CH933810 EDW06827.1 KRF93830.1 OUUW01000003 SPP78461.1 CH940664 EDW62920.1 KRF80855.1 UFQT01000258 SSX22498.1 AJVK01013239 CVRI01000014 CRK89755.1 DS235784 EEB16974.1 CH379063 EAL32672.2 ATLV01020054 KE525293 KFB44783.1 AJWK01026607 CH479204 EDW30549.1 LJIJ01000051 ODN04213.1 LNIX01000001 OXA65160.1 LNIX01000003 OXA57585.1 LJIJ01000363 ODM98332.1 BDOT01000024 GCF45299.1 JH817988 EKC30591.1 CM012455 RVE59572.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000053268
UP000007266
+ More
UP000037510 UP000008820 UP000075885 UP000002320 UP000075840 UP000075903 UP000002282 UP000075886 UP000000803 UP000008711 UP000000304 UP000075902 UP000076408 UP000007801 UP000007798 UP000001070 UP000245037 UP000075883 UP000075901 UP000075900 UP000092553 UP000075920 UP000009192 UP000268350 UP000008792 UP000075880 UP000075884 UP000092462 UP000183832 UP000075881 UP000009046 UP000076407 UP000001819 UP000030765 UP000092461 UP000008744 UP000094527 UP000198287 UP000079169 UP000261660 UP000288954 UP000005408
UP000037510 UP000008820 UP000075885 UP000002320 UP000075840 UP000075903 UP000002282 UP000075886 UP000000803 UP000008711 UP000000304 UP000075902 UP000076408 UP000007801 UP000007798 UP000001070 UP000245037 UP000075883 UP000075901 UP000075900 UP000092553 UP000075920 UP000009192 UP000268350 UP000008792 UP000075880 UP000075884 UP000092462 UP000183832 UP000075881 UP000009046 UP000076407 UP000001819 UP000030765 UP000092461 UP000008744 UP000094527 UP000198287 UP000079169 UP000261660 UP000288954 UP000005408
Interpro
Gene 3D
ProteinModelPortal
H9JTB4
A0A2A4K2J8
A0A437BFA1
A0A194RSH8
A0A194Q0U5
D2A4Z1
+ More
V5I996 A0A0T6B029 A0A0L7LRY8 A0A1L8DK35 A0A1L8DJT1 A0A1L8DJW2 Q17PW3 A0A023ESG1 A0A1S4HDU9 A0A1Q3FE42 A0A182PI93 B0W0M1 A0A182HFX7 A0A182VB23 B4PXI3 A0A182Q1B9 Q9V3S7 A0A0K8US37 B3NW34 B4R7J4 A0A034W2K1 A0A182TNH3 A0A182YEC0 B3MY19 B4MJB1 A0A0A1WQL9 B4JKQ9 A0A2P8Y1G1 A0A182M8H0 A0A182SYF7 A0A182RCZ3 A0A0M4EIN8 A0A182WDE2 B4L2F1 A0A3B0K0C8 B4MEE5 A0A182JJ18 A0A336M8X9 A0A182MXI7 A0A1B0DA96 A0A1J1HP75 A0A182JSX4 E0VUB8 A0A182X5H9 Q29II1 A0A084W3I9 A0A1B0CSU3 B4H2I2 A0A1D2NG18 A0A226F756 A0A1S3CVZ4 A0A226EIS5 A0A1D2MZ95 A0A3Q3EPE1 A0A402EMB0 K1R9G4 A0A437C9X4
V5I996 A0A0T6B029 A0A0L7LRY8 A0A1L8DK35 A0A1L8DJT1 A0A1L8DJW2 Q17PW3 A0A023ESG1 A0A1S4HDU9 A0A1Q3FE42 A0A182PI93 B0W0M1 A0A182HFX7 A0A182VB23 B4PXI3 A0A182Q1B9 Q9V3S7 A0A0K8US37 B3NW34 B4R7J4 A0A034W2K1 A0A182TNH3 A0A182YEC0 B3MY19 B4MJB1 A0A0A1WQL9 B4JKQ9 A0A2P8Y1G1 A0A182M8H0 A0A182SYF7 A0A182RCZ3 A0A0M4EIN8 A0A182WDE2 B4L2F1 A0A3B0K0C8 B4MEE5 A0A182JJ18 A0A336M8X9 A0A182MXI7 A0A1B0DA96 A0A1J1HP75 A0A182JSX4 E0VUB8 A0A182X5H9 Q29II1 A0A084W3I9 A0A1B0CSU3 B4H2I2 A0A1D2NG18 A0A226F756 A0A1S3CVZ4 A0A226EIS5 A0A1D2MZ95 A0A3Q3EPE1 A0A402EMB0 K1R9G4 A0A437C9X4
Ontologies
GO
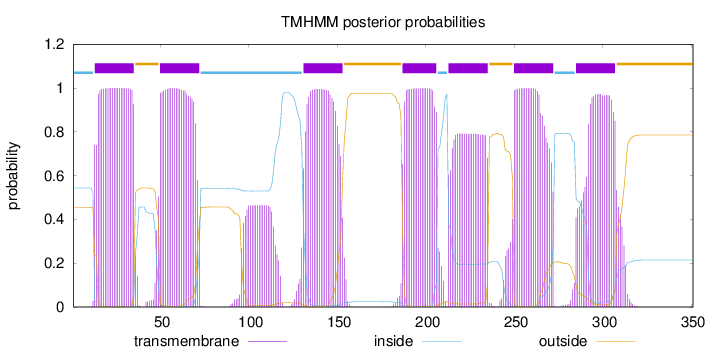
Topology
Length:
351
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
158.37534
Exp number, first 60 AAs:
34.76826
Total prob of N-in:
0.54386
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 35
outside
36 - 49
TMhelix
50 - 72
inside
73 - 130
TMhelix
131 - 153
outside
154 - 186
TMhelix
187 - 206
inside
207 - 212
TMhelix
213 - 235
outside
236 - 249
TMhelix
250 - 272
inside
273 - 284
TMhelix
285 - 307
outside
308 - 351
Population Genetic Test Statistics
Pi
264.723344
Theta
180.263957
Tajima's D
1.982814
CLR
0.180533
CSRT
0.878806059697015
Interpretation
Uncertain
