Gene
KWMTBOMO09116
Pre Gene Modal
BGIBMGA007534
Annotation
E2F_family_member_8_[Danaus_plexippus]
Full name
Transcription factor E2F7
Transcription factor
Location in the cell
Mitochondrial Reliability : 2.111 Nuclear Reliability : 1.931
Sequence
CDS
ATGTATGAAGATGATAACATCACTTGTACCCCAAAGCGTTTTGTGCTCACTGAAGTTACGAATTCATCGAGTTATGTATCGCCGACAGCAAATCTCAAATTACTAACAAATGTCGCGTTACAATATCCTACACCGCCGCCCAGTGCTGTGCAACGCAAAGAGAAATCACTTCAAATATTGTGTGACAGATTCTTACAGCTCTATCCACTCAAAGGTAATGGAACGGTGGAAATTCAGTTGGACAGTACTGCAGCCAGGTTAGGTGTCGAAAAGCGACGTATATACGACATCATTAACATCTTAGAAGCCATGCAATGTGCTGTTCATAAAAGGAAGAATACATATTTGTGGCATGGGGGATCAAGATTAAATTCCTTCTTGAAAATGTTAAAAAAACAAGGGGAGAGCATGAGACTGTCGGAGGCCCTCAGAGGTAAAGCACCAAAGCCTCCAGCGCCTAGACATAAAACATTAGGGGTCCTGGCTCAAAGATTCCTAATGTTGTTCTTAGTAGAACCTCTAAATACATTAATCAATTTAGAAATGGCTGTGAGGGTACTTATTGACACATCTAACAAGAACAAGACAAATCTCTCCCTGGAACAACAGGATCGGCAACATAAATCTAAAGTGAGGAGATTGTATGATATTGCCAATGTATTGATATCCATTGGTCTTATAGAAAAAGTATCCGGAAATTTAATATTAAAGAAACCTGTATTTAAATATGTGGGGCCTTGCAAAATGAAAAAATGTGAAAAAACAATGTTAACAACCCCATCCCCTATTACCCCTGTAACTGTATTGGAAAGCCAGTCTAAGACACCGACATTGCAAGTATTTGCAGGAAAATCTAAACGGAAATTAGAGTTCACTACTCCAAGTTCGACCAAAGAAATGAAACTTGGTATCACAACCCCACCTCACACTCCGTCACACAAATGGGATGAAATCCTGCTCGTAGCTGACATGGAACTGACTAGAATTAACAATGGAGCTGTTTTGTGA
Protein
MYEDDNITCTPKRFVLTEVTNSSSYVSPTANLKLLTNVALQYPTPPPSAVQRKEKSLQILCDRFLQLYPLKGNGTVEIQLDSTAARLGVEKRRIYDIINILEAMQCAVHKRKNTYLWHGGSRLNSFLKMLKKQGESMRLSEALRGKAPKPPAPRHKTLGVLAQRFLMLFLVEPLNTLINLEMAVRVLIDTSNKNKTNLSLEQQDRQHKSKVRRLYDIANVLISIGLIEKVSGNLILKKPVFKYVGPCKMKKCEKTMLTTPSPITPVTVLESQSKTPTLQVFAGKSKRKLEFTTPSSTKEMKLGITTPPHTPSHKWDEILLVADMELTRINNGAVL
Summary
Description
Atypical E2F transcription factor that participates in various processes such as angiogenesis, polyploidization of specialized cells and DNA damage response. Mainly acts as a transcription repressor that binds DNA independently of DP proteins and specifically recognizes the E2 recognition site 5'-TTTC[CG]CGC-3'. Directly represses transcription of classical E2F transcription factors such as E2F1. Acts as a regulator of S-phase by recognizing and binding the E2-related site 5'-TTCCCGCC-3' and mediating repression of G1/S-regulated genes. Plays a key role in polyploidization of cells in placenta and liver by regulating the endocycle, probably by repressing genes promoting cytokinesis and antagonizing action of classical E2F proteins (E2F1, E2F2 and/or E2F3). Required for placental development by promoting polyploidization of trophoblast giant cells. Also involved in DNA damage response: up-regulated by p53/TP53 following genotoxic stress and acts as a downstream effector of p53/TP53-dependent repression by mediating repression of indirect p53/TP53 target genes involved in DNA replication. Acts as a promoter of sprouting angiogenesis, possibly by acting as a transcription activator: associates with HIF1A, recognizes and binds the VEGFA promoter, which is different from canonical E2 recognition site, and activates expression of the VEGFA gene. Acts as a negative regulator of keratinocyte differentiation.
Subunit
Interacts with HIF1A (By similarity). Homodimer and heterodimer: mainly forms homodimers and, to a lesser extent, heterodimers with E2F8. Dimerization is important for DNA-binding.
Similarity
Belongs to the E2F/DP family.
Keywords
Activator
Alternative splicing
Cell cycle
Complete proteome
DNA damage
DNA-binding
Nucleus
Phosphoprotein
Reference proteome
Repressor
Transcription
Transcription regulation
Feature
chain Transcription factor E2F7
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9JDD7
A0A2H1VMW2
A0A2A4JKR3
A0A212FCN2
A0A0L7LA70
A0A437BGG8
+ More
A0A194R7U2 A0A194PL59 K7J4J1 A0A232ENG1 A0A2P8YM49 A0A1B6KZI0 A0A1B6FKL6 F6XM19 A0A1W5BDL8 A0A1W5BHR1 A0A1B6DN63 A0A087XBM3 M3ZVM7 A0A3B4BAK2 A0A3Q3N551 A0A3B3XJ21 A0A3P9PFU2 A0A3Q2VIM8 A0A3P9BNN3 A0A3P8QA10 A0A3Q3QPF2 A0A2I4AIZ9 A0A315USB9 A0A3Q3E5C7 A0A3Q1GCK6 A0A401NS85 B7QG98 A0A329SYE1 A0A3Q1CGF1 Q6NRA1 D8SRS2 A0A1L8GIF4 Q32NV0 A0A3B4X493 A0A3B4UWY3 A0A329SYR9 H2RVT7 A0A1V9ZA47 T2M8C1 Q6S7F2-2 Q6S7F2 A0A093BRA0
A0A194R7U2 A0A194PL59 K7J4J1 A0A232ENG1 A0A2P8YM49 A0A1B6KZI0 A0A1B6FKL6 F6XM19 A0A1W5BDL8 A0A1W5BHR1 A0A1B6DN63 A0A087XBM3 M3ZVM7 A0A3B4BAK2 A0A3Q3N551 A0A3B3XJ21 A0A3P9PFU2 A0A3Q2VIM8 A0A3P9BNN3 A0A3P8QA10 A0A3Q3QPF2 A0A2I4AIZ9 A0A315USB9 A0A3Q3E5C7 A0A3Q1GCK6 A0A401NS85 B7QG98 A0A329SYE1 A0A3Q1CGF1 Q6NRA1 D8SRS2 A0A1L8GIF4 Q32NV0 A0A3B4X493 A0A3B4UWY3 A0A329SYR9 H2RVT7 A0A1V9ZA47 T2M8C1 Q6S7F2-2 Q6S7F2 A0A093BRA0
Pubmed
EMBL
BABH01030090
ODYU01003413
SOQ42148.1
NWSH01001151
PCG72376.1
AGBW02009176
+ More
OWR51468.1 JTDY01001989 KOB72388.1 RSAL01000065 RVE49438.1 KQ460615 KPJ13717.1 KQ459602 KPI93469.1 AAZX01009232 NNAY01003159 OXU19890.1 PYGN01000498 PSN45327.1 GEBQ01023114 JAT16863.1 GECZ01019044 JAS50725.1 EAAA01001229 GEDC01010898 GEDC01010190 JAS26400.1 JAS27108.1 AYCK01014070 NHOQ01002778 PWA14695.1 BFAA01005383 GCB63758.1 ABJB010103991 ABJB010692593 ABJB010719844 ABJB011002344 ABJB011072064 DS930038 EEC17870.1 MJFZ01000025 RAW41734.1 BC070864 AAH70864.1 GL377636 EFJ12836.1 CM004472 OCT83600.1 BC108466 AAI08467.1 RAW41735.1 JNBS01002161 OQR94874.1 HAAD01001990 CDG68222.1 AY463359 AK031078 AK045040 AK041481 AK143686 CH466539 BC145429 BC150772 KL229496 KFV04718.1
OWR51468.1 JTDY01001989 KOB72388.1 RSAL01000065 RVE49438.1 KQ460615 KPJ13717.1 KQ459602 KPI93469.1 AAZX01009232 NNAY01003159 OXU19890.1 PYGN01000498 PSN45327.1 GEBQ01023114 JAT16863.1 GECZ01019044 JAS50725.1 EAAA01001229 GEDC01010898 GEDC01010190 JAS26400.1 JAS27108.1 AYCK01014070 NHOQ01002778 PWA14695.1 BFAA01005383 GCB63758.1 ABJB010103991 ABJB010692593 ABJB010719844 ABJB011002344 ABJB011072064 DS930038 EEC17870.1 MJFZ01000025 RAW41734.1 BC070864 AAH70864.1 GL377636 EFJ12836.1 CM004472 OCT83600.1 BC108466 AAI08467.1 RAW41735.1 JNBS01002161 OQR94874.1 HAAD01001990 CDG68222.1 AY463359 AK031078 AK045040 AK041481 AK143686 CH466539 BC145429 BC150772 KL229496 KFV04718.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000037510
UP000283053
UP000053240
+ More
UP000053268 UP000002358 UP000215335 UP000245037 UP000008144 UP000028760 UP000002852 UP000261520 UP000261640 UP000261480 UP000242638 UP000264840 UP000265160 UP000265100 UP000261600 UP000192220 UP000261660 UP000257200 UP000288216 UP000001555 UP000251314 UP000257160 UP000001514 UP000186698 UP000261360 UP000261420 UP000005226 UP000243217 UP000000589
UP000053268 UP000002358 UP000215335 UP000245037 UP000008144 UP000028760 UP000002852 UP000261520 UP000261640 UP000261480 UP000242638 UP000264840 UP000265160 UP000265100 UP000261600 UP000192220 UP000261660 UP000257200 UP000288216 UP000001555 UP000251314 UP000257160 UP000001514 UP000186698 UP000261360 UP000261420 UP000005226 UP000243217 UP000000589
Pfam
PF02319 E2F_TDP
Interpro
SUPFAM
SSF46785
SSF46785
Gene 3D
ProteinModelPortal
H9JDD7
A0A2H1VMW2
A0A2A4JKR3
A0A212FCN2
A0A0L7LA70
A0A437BGG8
+ More
A0A194R7U2 A0A194PL59 K7J4J1 A0A232ENG1 A0A2P8YM49 A0A1B6KZI0 A0A1B6FKL6 F6XM19 A0A1W5BDL8 A0A1W5BHR1 A0A1B6DN63 A0A087XBM3 M3ZVM7 A0A3B4BAK2 A0A3Q3N551 A0A3B3XJ21 A0A3P9PFU2 A0A3Q2VIM8 A0A3P9BNN3 A0A3P8QA10 A0A3Q3QPF2 A0A2I4AIZ9 A0A315USB9 A0A3Q3E5C7 A0A3Q1GCK6 A0A401NS85 B7QG98 A0A329SYE1 A0A3Q1CGF1 Q6NRA1 D8SRS2 A0A1L8GIF4 Q32NV0 A0A3B4X493 A0A3B4UWY3 A0A329SYR9 H2RVT7 A0A1V9ZA47 T2M8C1 Q6S7F2-2 Q6S7F2 A0A093BRA0
A0A194R7U2 A0A194PL59 K7J4J1 A0A232ENG1 A0A2P8YM49 A0A1B6KZI0 A0A1B6FKL6 F6XM19 A0A1W5BDL8 A0A1W5BHR1 A0A1B6DN63 A0A087XBM3 M3ZVM7 A0A3B4BAK2 A0A3Q3N551 A0A3B3XJ21 A0A3P9PFU2 A0A3Q2VIM8 A0A3P9BNN3 A0A3P8QA10 A0A3Q3QPF2 A0A2I4AIZ9 A0A315USB9 A0A3Q3E5C7 A0A3Q1GCK6 A0A401NS85 B7QG98 A0A329SYE1 A0A3Q1CGF1 Q6NRA1 D8SRS2 A0A1L8GIF4 Q32NV0 A0A3B4X493 A0A3B4UWY3 A0A329SYR9 H2RVT7 A0A1V9ZA47 T2M8C1 Q6S7F2-2 Q6S7F2 A0A093BRA0
PDB
4YO2
E-value=2.84855e-23,
Score=268
Ontologies
GO
GO:0005634
GO:0003677
GO:0005667
GO:0003700
GO:0043565
GO:0051726
GO:0000981
GO:0008134
GO:0000122
GO:0090575
GO:0045944
GO:0001946
GO:0008045
GO:0045892
GO:0002040
GO:0000978
GO:0032466
GO:0060707
GO:0006355
GO:0000987
GO:0060718
GO:2000134
GO:0030330
GO:0003714
GO:0001890
GO:0008285
GO:0042802
GO:0071930
GO:0016607
GO:0032877
GO:0070365
GO:0001227
GO:0000977
GO:0019205
GO:0005739
GO:0016776
GO:0006811
GO:0016020
GO:0016876
GO:0043039
PANTHER
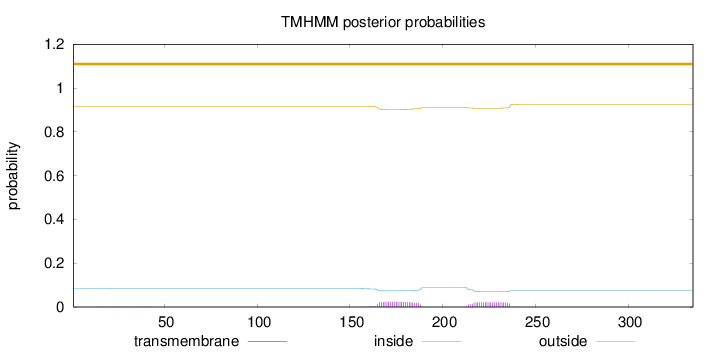
Topology
Subcellular location
Nucleus
Length:
335
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.988089999999999
Exp number, first 60 AAs:
0.0088
Total prob of N-in:
0.08447
outside
1 - 335
Population Genetic Test Statistics
Pi
29.672524
Theta
56.795949
Tajima's D
-2.13712
CLR
4979.149245
CSRT
0.00864956752162392
Interpretation
Uncertain
