Gene
KWMTBOMO08958
Pre Gene Modal
BGIBMGA007797
Annotation
PREDICTED:_zinc-responsive_transcriptional_regulator_ZAP1-like_[Papilio_polytes]
Transcription factor
Location in the cell
Nuclear Reliability : 4.282
Sequence
CDS
ATGCGATCTGTAAACGTGTTCATGGAATCGCCCGACTCTAGGTCGAATCCTTTGTTATCGGAGGAACCGAAAGAACAGAACAACAATAGCTTGTTAAGTGTTGCGACGACGCAGGCGAACACAAGCGCAATGACAGCCGCGACTACTTCTACTACACTACAAAGTTTCGACTCGATATGGAATGTAGATCGTGATCGAGATCGCTTGGACACTTCAATGCTAGAGGACTTAAATAAATTTAGTTATTGGAACCAAGATAGTGAAATAGCGGACACCCCCTGCACGGATACGACAATTTCAAATAAACTAATCAACAATACCGATGGTCAAATATATACTCTTACTGTCCTCAATCACGATATAACAGAAAACAGTTCAAGTCGCTATTGGAGTAAAGACGAAGACGTTTCTATGTCTAGTCCGGCAGACATAGAACCCAATCCTGCATTGGATCTGGAATCAATATTAAATATGAATGGATTCCCAAACGATTTCAACCAAGATCTCCCCAAACCTTTTCATTTTGAAGATGGTGATTCAGAGAGTCAAAACAATGATCTGAACTCAAGTAATCTAGTAAAAGTGGAACAATATTACGATGAAAACGAATATACAAACAGTGATAAGAATGATGACACGCAAAGCTCCTCTTTAATTGGTGCTGCAACTTTACTGGATACTCAAATCGAATTTAACAACAATAACAATGACTGGAAAGCAACAAATAACAATGCGAATGAATCTTTACTTAGAAGTGCATTACAGGGAAAAGCTTATTTCCGTACTGTTCAAAAAAGTACTGTTACTAAAATAGACGCCAATGCTGAATTGAAGCGAGTCATAATCAATAATAATAACAAAGCTGACGTACCATCTATGTACCAAGATGCAAAGGACGCCGATCCGGAATTATTGATGGCAATTTCACCTCCAAACGGAAATGTCAACATTTCAATTTTATTAGAGGATCCAAGCACAGCTGTTATAACTAATGGAGAAAACCCGACATCGACGCAGCAAAGCGTGGATGACATTTTGCTGTCTCAGTTGGATGCGAACTATCCTGAAGACTATGAAAAATTAAAAAGAATAGCCACGGAGCTTGGTGAATCAATGCAACCATTCTGCACTGTAGAACCCATTGATCCTACTCGAAGTGTCTACAACATCCATCATGTGAATGGTGAATTAGTGACGATGATTCCAGCTGGGGAAGTACAACTCCCTCATAACTTGCAAATAGTCACCGCTTCACCAGCAGTTACGAGCACAAAAAACACAAAGAGATCTCTGCATTACTGTTCTATTTGTTCAAAAGGGTTCAAAGATAAATATTCTGTAAACGTACACGTAAGAACACATACTGGGGAGAAACCGTTTACATGTTCCCTATGCGGAAAGAGTTTCAGACAGAAAGCACATCTCGCGAAGCACTACCAGACACACGTAGCACAAAAAGCGGCCGCCGCCAACGGCGCTGTCAAACCCAACAAGCAAAGGTAG
Protein
MRSVNVFMESPDSRSNPLLSEEPKEQNNNSLLSVATTQANTSAMTAATTSTTLQSFDSIWNVDRDRDRLDTSMLEDLNKFSYWNQDSEIADTPCTDTTISNKLINNTDGQIYTLTVLNHDITENSSSRYWSKDEDVSMSSPADIEPNPALDLESILNMNGFPNDFNQDLPKPFHFEDGDSESQNNDLNSSNLVKVEQYYDENEYTNSDKNDDTQSSSLIGAATLLDTQIEFNNNNNDWKATNNNANESLLRSALQGKAYFRTVQKSTVTKIDANAELKRVIINNNNKADVPSMYQDAKDADPELLMAISPPNGNVNISILLEDPSTAVITNGENPTSTQQSVDDILLSQLDANYPEDYEKLKRIATELGESMQPFCTVEPIDPTRSVYNIHHVNGELVTMIPAGEVQLPHNLQIVTASPAVTSTKNTKRSLHYCSICSKGFKDKYSVNVHVRTHTGEKPFTCSLCGKSFRQKAHLAKHYQTHVAQKAAAANGAVKPNKQR
Summary
Uniprot
Pubmed
EMBL
BABH01002596
ODYU01007308
SOQ49973.1
KQ459602
KPI93313.1
NWSH01000740
+ More
PCG74492.1 KQ460615 KPJ13557.1 AGBW02011580 OWR46461.1 JTDY01000315 KOB77733.1 KQ971342 EFA03748.1 GEZM01046801 GEZM01046800 JAV77001.1 LJIG01002645 KRT84295.1 PYGN01000056 PSN55996.1 KB632335 ERL92807.1 APGK01026260 APGK01026261 KB740624 ENN79949.1 NEVH01020964 PNF20289.1 AJWK01006634 AJWK01006635 AJWK01006636 AJWK01006637 AJWK01006638 GBHO01008316 GBHO01008315 GDHC01015803 JAG35288.1 JAG35289.1 JAQ02826.1 KK854096 PTY11191.1 KK854399 PTY15460.1 KQ414663 KOC65458.1
PCG74492.1 KQ460615 KPJ13557.1 AGBW02011580 OWR46461.1 JTDY01000315 KOB77733.1 KQ971342 EFA03748.1 GEZM01046801 GEZM01046800 JAV77001.1 LJIG01002645 KRT84295.1 PYGN01000056 PSN55996.1 KB632335 ERL92807.1 APGK01026260 APGK01026261 KB740624 ENN79949.1 NEVH01020964 PNF20289.1 AJWK01006634 AJWK01006635 AJWK01006636 AJWK01006637 AJWK01006638 GBHO01008316 GBHO01008315 GDHC01015803 JAG35288.1 JAG35289.1 JAQ02826.1 KK854096 PTY11191.1 KK854399 PTY15460.1 KQ414663 KOC65458.1
Proteomes
PRIDE
Pfam
PF00096 zf-C2H2
SUPFAM
SSF57667
SSF57667
ProteinModelPortal
PDB
5V3G
E-value=8.61246e-12,
Score=171
Ontologies
GO
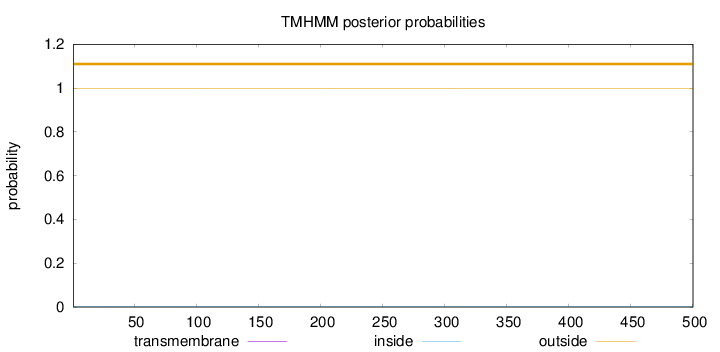
Topology
Length:
500
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0014
Exp number, first 60 AAs:
0.00133
Total prob of N-in:
0.00310
outside
1 - 500
Population Genetic Test Statistics
Pi
301.850822
Theta
233.89778
Tajima's D
1.073784
CLR
0.063379
CSRT
0.67631618419079
Interpretation
Uncertain
