Gene
KWMTBOMO08821
Pre Gene Modal
BGIBMGA007867
Annotation
PREDICTED:_zinc_finger_protein_665_isoform_X2_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 3.877
Sequence
CDS
ATGTTTACAACTAACAACGTTCAAGCGGGCACAGTGCCCACTCTACAGTACCAAGAACATCCTCAGAAGTCACAAGGGAAGAACATAGAAAACGTACAACAAAAGAATGCTCATCAATCACAGGCACAACAGGAATTTCCAACATTTTGTTATACAACAAATGTGAATATGATTGGTAAAATTGGAACTGCGAACACTAGTCAAACAGGGGGTGTGAACATAACGCAACTGACTACAAGCGATGACAAGACATGCTATATAGCACAACCGTTCTCATATAATTATGCACTTGTTAATCAAATGCAAATAGCTCCTAATGGAATACAAAATACAATATCAAACATTAGTTTCAAATGTGATGTTTGTGGACTTATGTTTGGTCATCTAACTTTACTGACAGCTCATAAAAGGATTCATACTCAAGATACAGATAACAATATTACAGTGGTAGCAACAGCTGGAAATGATGTTACAATGCCTTCACATATACAAATATTGTCATCAGACCCAAATGAACAACAAAATCAACAACACCATGTACAAATACAAGACACCAAGCCTTTACTAGACAAAATTCAGAAATGCATATCATGTGGTGGAACAATACAGAACAATCCAAAAAGAAAGGGGCCCAAGTTAATAAGATGTGAAACCTGTATAGCACAAGACTCTGTTGATCAACATAGACCAAATCAAATGATTCCGGCACAAATTTTCGTTGGAGAAGAGAATGTGAAGTATGAAGTTAGTGGTGTGAACTCTAATGCACCTTCAACAGATCCACTTTCGACCACTCAAACAGCTCAACCTGTGCAACAACAACAACAACAGCAGCAGCAACAGCAGCAAAAACCATTGCCAGGTCACCATCCTGTGAAAAAAAGAAATTTGTCATCTGTAACAAAATGTCAGAATTGCAATGGTTCTGGTATTATATTTGTCGGCGGAAACAAAAATAAGTTGAATCAGCCAAGTGCAGATAAACCTTTCCATTGTAACATTTGTGGGGGCTCATTTTCGAGATATTCTTCGTTGTGGTCCCACAAGAAATTACATTCTGGTGAGAAGAATTTTAAATGCGGTATTTGTGGCATAGCTTTCGCAAAAGCTGTTTACCTTAAAAACCATTCAAGAATACACACTGGAGAGAAACCTTACCGGTGTAAAACTTGTGGAATGCAGTTTTCACAATCTCCTCATCTAAAAAACCATGAAAGAACACATTCTGGAGAAAAACCCTATGTTTGTGAGGTTTGCGACAAAGGCTTTGCGCGACACGCAACGCTCTGGAACCATCGACGCATACACACTGGAGAGAAACCATACAAATGTGAAACTTGCGGATCAGCATTCAGCCAAGCTGCTCACTTAAAGAATCACGCGAAAGTACATTCAGGAGAGAAACCTTTCAAATGTGATATTTGTACGGCAGCCTTCGCGGATCGGTTTGCCTTGAAGAGACATAGAGGAATACATGATAAATACGGTCAAACTGCGCCTTTACCTTCGAGTATACATTTAGATCGGCAACAAAATCAAACTCTTCAGCCATCTCAGCAATCCAACAACGAACAAAGCACAACACAAACCAGTGTTGAAGTAGTCGTGACGCCACAGGGAACGACAACTGTCAGCATAGGGAATTGA
Protein
MFTTNNVQAGTVPTLQYQEHPQKSQGKNIENVQQKNAHQSQAQQEFPTFCYTTNVNMIGKIGTANTSQTGGVNITQLTTSDDKTCYIAQPFSYNYALVNQMQIAPNGIQNTISNISFKCDVCGLMFGHLTLLTAHKRIHTQDTDNNITVVATAGNDVTMPSHIQILSSDPNEQQNQQHHVQIQDTKPLLDKIQKCISCGGTIQNNPKRKGPKLIRCETCIAQDSVDQHRPNQMIPAQIFVGEENVKYEVSGVNSNAPSTDPLSTTQTAQPVQQQQQQQQQQQQKPLPGHHPVKKRNLSSVTKCQNCNGSGIIFVGGNKNKLNQPSADKPFHCNICGGSFSRYSSLWSHKKLHSGEKNFKCGICGIAFAKAVYLKNHSRIHTGEKPYRCKTCGMQFSQSPHLKNHERTHSGEKPYVCEVCDKGFARHATLWNHRRIHTGEKPYKCETCGSAFSQAAHLKNHAKVHSGEKPFKCDICTAAFADRFALKRHRGIHDKYGQTAPLPSSIHLDRQQNQTLQPSQQSNNEQSTTQTSVEVVVTPQGTTTVSIGN
Summary
Uniprot
H9JEC0
A0A2H1VLE2
A0A3S2TE07
A0A212FEE5
A0A194QMF3
S4PFV9
+ More
A0A194PWM2 A0A2A4J6B6 A0A1L8DJ68 A0A0K8TP08 A0A2W1B9Y0 B0WIT6 U5EQX0 Q16JY2 A0A084VT27 A0A2M4BIJ3 A0A2M3Z1C4 A0A2M3Z1I7 A0A1L8DJ08 A0A2M4AA15 A0A2M4CQB8 W5JT53 A0A2M4BKM2 A0A1W4XDQ2 A0A0L7KXD8 A0A1W4XNB8 B4IIJ6 A0A0Q9WJR8 D2A665 A0A0Q9X4N8 B3N1C7 B3NKN7 Q9V9N4 B4P6D4 A0A0P8XII9 B4MUP1 B4LQS8 B4JBM4 Q8IGP5 A0A0Q5VVY1 B4KFV7 A0A0R1DP70 B4GKV3 Q29K11 A0A3B0JYN7 A0A1W4W849 A0A1W4W8L7 A0A0R3NVJ6 A0A3B0JVV0 A0A0M4E4Q5 A0A0J9R5Q5 A0A0J9R5E3 A0A1I8MZF2 Q961A1 N6U0Q6 Q7PWW4 A0A3B0JVQ5 A0A1B0FLC5 A0A0T6BCI4 A0A0M8ZZ65 A0A146MD80 A0A0A9YYS1 A0A2A3EEP8 A0A0C9QUE7 A0A0J7NZD0 A0A069DZ60 A0A0V0GAV0 A0A023F276 E2A270 A0A2R7WQC2 A0A336MSL6 A0A0C9RQQ0 A0A3L8E0K2 A0A026WE63 A0A151WEM6 F4X371 A0A158NM94 A0A232FHY5 A0A151IVY5 A0A195FHW7 A0A195BER4 A0A2S2R7B8
A0A194PWM2 A0A2A4J6B6 A0A1L8DJ68 A0A0K8TP08 A0A2W1B9Y0 B0WIT6 U5EQX0 Q16JY2 A0A084VT27 A0A2M4BIJ3 A0A2M3Z1C4 A0A2M3Z1I7 A0A1L8DJ08 A0A2M4AA15 A0A2M4CQB8 W5JT53 A0A2M4BKM2 A0A1W4XDQ2 A0A0L7KXD8 A0A1W4XNB8 B4IIJ6 A0A0Q9WJR8 D2A665 A0A0Q9X4N8 B3N1C7 B3NKN7 Q9V9N4 B4P6D4 A0A0P8XII9 B4MUP1 B4LQS8 B4JBM4 Q8IGP5 A0A0Q5VVY1 B4KFV7 A0A0R1DP70 B4GKV3 Q29K11 A0A3B0JYN7 A0A1W4W849 A0A1W4W8L7 A0A0R3NVJ6 A0A3B0JVV0 A0A0M4E4Q5 A0A0J9R5Q5 A0A0J9R5E3 A0A1I8MZF2 Q961A1 N6U0Q6 Q7PWW4 A0A3B0JVQ5 A0A1B0FLC5 A0A0T6BCI4 A0A0M8ZZ65 A0A146MD80 A0A0A9YYS1 A0A2A3EEP8 A0A0C9QUE7 A0A0J7NZD0 A0A069DZ60 A0A0V0GAV0 A0A023F276 E2A270 A0A2R7WQC2 A0A336MSL6 A0A0C9RQQ0 A0A3L8E0K2 A0A026WE63 A0A151WEM6 F4X371 A0A158NM94 A0A232FHY5 A0A151IVY5 A0A195FHW7 A0A195BER4 A0A2S2R7B8
Pubmed
19121390
22118469
26354079
23622113
26369729
28756777
+ More
17510324 24438588 20920257 23761445 26227816 17994087 18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 18057021 15632085 23185243 22936249 25315136 23537049 12364791 26823975 25401762 26334808 25474469 20798317 30249741 24508170 21719571 21347285 28648823
17510324 24438588 20920257 23761445 26227816 17994087 18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 18057021 15632085 23185243 22936249 25315136 23537049 12364791 26823975 25401762 26334808 25474469 20798317 30249741 24508170 21719571 21347285 28648823
EMBL
BABH01002864
BABH01002865
ODYU01003012
SOQ41242.1
RSAL01000266
RVE43272.1
+ More
AGBW02008937 OWR52126.1 KQ461195 KPJ06723.1 GAIX01003867 JAA88693.1 KQ459596 KPI95535.1 NWSH01002960 PCG67218.1 GFDF01007674 JAV06410.1 GDAI01001973 JAI15630.1 KZ150372 PZC71135.1 DS231952 EDS28749.1 GANO01004049 JAB55822.1 CH477986 EAT34601.1 ATLV01016154 KE525057 KFB41121.1 GGFJ01003693 MBW52834.1 GGFM01001487 MBW22238.1 GGFM01001646 MBW22397.1 GFDF01007729 JAV06355.1 GGFK01004137 MBW37458.1 GGFL01003354 MBW67532.1 ADMH02000519 ETN66125.1 GGFJ01004372 MBW53513.1 JTDY01004756 KOB67795.1 CH480843 EDW49746.1 CH940649 KRF81131.1 KQ971346 EFA04970.2 CH933807 KRG03078.1 CH902652 EDV33648.1 CH954179 EDV54341.1 AE014134 BT001891 AAF57252.1 AAN71680.1 CM000158 EDW89891.1 KPU74638.1 KPU74639.1 CH963857 EDW76236.1 EDW63462.1 CH916368 EDW02959.1 BT001676 AAN71431.1 AAX52677.1 KQS61774.1 KQS61775.1 EDW12083.1 KRJ98766.1 CH479184 EDW37269.1 CH379062 EAL32800.1 OUUW01000010 SPP86183.1 KRT04971.1 KRT04972.1 SPP86184.1 CP012523 ALC38921.1 CM002910 KMY91423.1 KMY91422.1 AY051742 AAK93166.1 APGK01044227 KB741021 ENN75090.1 AAAB01008984 EAA14852.5 SPP86185.1 CCAG010022749 LJIG01001890 KRT85050.1 KQ435803 KOX73076.1 GDHC01009991 GDHC01005358 GDHC01000935 JAQ08638.1 JAQ13271.1 JAQ17694.1 GBHO01007356 GBHO01001187 GBHO01001184 JAG36248.1 JAG42417.1 JAG42420.1 KZ288266 PBC30263.1 GBYB01007379 JAG77146.1 LBMM01000687 KMQ97755.1 GBGD01001205 JAC87684.1 GECL01001735 JAP04389.1 GBBI01003020 JAC15692.1 GL435922 EFN72475.1 KK855222 PTY21511.1 UFQS01002467 UFQT01002467 SSX14118.1 SSX33534.1 GBYB01009571 JAG79338.1 QOIP01000002 RLU25548.1 KK107250 EZA54395.1 KQ983238 KYQ46256.1 GL888609 EGI59052.1 ADTU01000293 NNAY01000169 OXU30351.1 KQ980895 KYN11755.1 KQ981606 KYN39584.1 KQ976509 KYM82677.1 GGMS01016692 MBY85895.1
AGBW02008937 OWR52126.1 KQ461195 KPJ06723.1 GAIX01003867 JAA88693.1 KQ459596 KPI95535.1 NWSH01002960 PCG67218.1 GFDF01007674 JAV06410.1 GDAI01001973 JAI15630.1 KZ150372 PZC71135.1 DS231952 EDS28749.1 GANO01004049 JAB55822.1 CH477986 EAT34601.1 ATLV01016154 KE525057 KFB41121.1 GGFJ01003693 MBW52834.1 GGFM01001487 MBW22238.1 GGFM01001646 MBW22397.1 GFDF01007729 JAV06355.1 GGFK01004137 MBW37458.1 GGFL01003354 MBW67532.1 ADMH02000519 ETN66125.1 GGFJ01004372 MBW53513.1 JTDY01004756 KOB67795.1 CH480843 EDW49746.1 CH940649 KRF81131.1 KQ971346 EFA04970.2 CH933807 KRG03078.1 CH902652 EDV33648.1 CH954179 EDV54341.1 AE014134 BT001891 AAF57252.1 AAN71680.1 CM000158 EDW89891.1 KPU74638.1 KPU74639.1 CH963857 EDW76236.1 EDW63462.1 CH916368 EDW02959.1 BT001676 AAN71431.1 AAX52677.1 KQS61774.1 KQS61775.1 EDW12083.1 KRJ98766.1 CH479184 EDW37269.1 CH379062 EAL32800.1 OUUW01000010 SPP86183.1 KRT04971.1 KRT04972.1 SPP86184.1 CP012523 ALC38921.1 CM002910 KMY91423.1 KMY91422.1 AY051742 AAK93166.1 APGK01044227 KB741021 ENN75090.1 AAAB01008984 EAA14852.5 SPP86185.1 CCAG010022749 LJIG01001890 KRT85050.1 KQ435803 KOX73076.1 GDHC01009991 GDHC01005358 GDHC01000935 JAQ08638.1 JAQ13271.1 JAQ17694.1 GBHO01007356 GBHO01001187 GBHO01001184 JAG36248.1 JAG42417.1 JAG42420.1 KZ288266 PBC30263.1 GBYB01007379 JAG77146.1 LBMM01000687 KMQ97755.1 GBGD01001205 JAC87684.1 GECL01001735 JAP04389.1 GBBI01003020 JAC15692.1 GL435922 EFN72475.1 KK855222 PTY21511.1 UFQS01002467 UFQT01002467 SSX14118.1 SSX33534.1 GBYB01009571 JAG79338.1 QOIP01000002 RLU25548.1 KK107250 EZA54395.1 KQ983238 KYQ46256.1 GL888609 EGI59052.1 ADTU01000293 NNAY01000169 OXU30351.1 KQ980895 KYN11755.1 KQ981606 KYN39584.1 KQ976509 KYM82677.1 GGMS01016692 MBY85895.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053240
UP000053268
UP000218220
+ More
UP000002320 UP000008820 UP000030765 UP000000673 UP000192223 UP000037510 UP000001292 UP000008792 UP000007266 UP000009192 UP000007801 UP000008711 UP000000803 UP000002282 UP000007798 UP000001070 UP000008744 UP000001819 UP000268350 UP000192221 UP000092553 UP000095301 UP000019118 UP000007062 UP000092444 UP000053105 UP000242457 UP000036403 UP000000311 UP000279307 UP000053097 UP000075809 UP000007755 UP000005205 UP000215335 UP000078492 UP000078541 UP000078540
UP000002320 UP000008820 UP000030765 UP000000673 UP000192223 UP000037510 UP000001292 UP000008792 UP000007266 UP000009192 UP000007801 UP000008711 UP000000803 UP000002282 UP000007798 UP000001070 UP000008744 UP000001819 UP000268350 UP000192221 UP000092553 UP000095301 UP000019118 UP000007062 UP000092444 UP000053105 UP000242457 UP000036403 UP000000311 UP000279307 UP000053097 UP000075809 UP000007755 UP000005205 UP000215335 UP000078492 UP000078541 UP000078540
Pfam
PF00096 zf-C2H2
SUPFAM
SSF57667
SSF57667
ProteinModelPortal
H9JEC0
A0A2H1VLE2
A0A3S2TE07
A0A212FEE5
A0A194QMF3
S4PFV9
+ More
A0A194PWM2 A0A2A4J6B6 A0A1L8DJ68 A0A0K8TP08 A0A2W1B9Y0 B0WIT6 U5EQX0 Q16JY2 A0A084VT27 A0A2M4BIJ3 A0A2M3Z1C4 A0A2M3Z1I7 A0A1L8DJ08 A0A2M4AA15 A0A2M4CQB8 W5JT53 A0A2M4BKM2 A0A1W4XDQ2 A0A0L7KXD8 A0A1W4XNB8 B4IIJ6 A0A0Q9WJR8 D2A665 A0A0Q9X4N8 B3N1C7 B3NKN7 Q9V9N4 B4P6D4 A0A0P8XII9 B4MUP1 B4LQS8 B4JBM4 Q8IGP5 A0A0Q5VVY1 B4KFV7 A0A0R1DP70 B4GKV3 Q29K11 A0A3B0JYN7 A0A1W4W849 A0A1W4W8L7 A0A0R3NVJ6 A0A3B0JVV0 A0A0M4E4Q5 A0A0J9R5Q5 A0A0J9R5E3 A0A1I8MZF2 Q961A1 N6U0Q6 Q7PWW4 A0A3B0JVQ5 A0A1B0FLC5 A0A0T6BCI4 A0A0M8ZZ65 A0A146MD80 A0A0A9YYS1 A0A2A3EEP8 A0A0C9QUE7 A0A0J7NZD0 A0A069DZ60 A0A0V0GAV0 A0A023F276 E2A270 A0A2R7WQC2 A0A336MSL6 A0A0C9RQQ0 A0A3L8E0K2 A0A026WE63 A0A151WEM6 F4X371 A0A158NM94 A0A232FHY5 A0A151IVY5 A0A195FHW7 A0A195BER4 A0A2S2R7B8
A0A194PWM2 A0A2A4J6B6 A0A1L8DJ68 A0A0K8TP08 A0A2W1B9Y0 B0WIT6 U5EQX0 Q16JY2 A0A084VT27 A0A2M4BIJ3 A0A2M3Z1C4 A0A2M3Z1I7 A0A1L8DJ08 A0A2M4AA15 A0A2M4CQB8 W5JT53 A0A2M4BKM2 A0A1W4XDQ2 A0A0L7KXD8 A0A1W4XNB8 B4IIJ6 A0A0Q9WJR8 D2A665 A0A0Q9X4N8 B3N1C7 B3NKN7 Q9V9N4 B4P6D4 A0A0P8XII9 B4MUP1 B4LQS8 B4JBM4 Q8IGP5 A0A0Q5VVY1 B4KFV7 A0A0R1DP70 B4GKV3 Q29K11 A0A3B0JYN7 A0A1W4W849 A0A1W4W8L7 A0A0R3NVJ6 A0A3B0JVV0 A0A0M4E4Q5 A0A0J9R5Q5 A0A0J9R5E3 A0A1I8MZF2 Q961A1 N6U0Q6 Q7PWW4 A0A3B0JVQ5 A0A1B0FLC5 A0A0T6BCI4 A0A0M8ZZ65 A0A146MD80 A0A0A9YYS1 A0A2A3EEP8 A0A0C9QUE7 A0A0J7NZD0 A0A069DZ60 A0A0V0GAV0 A0A023F276 E2A270 A0A2R7WQC2 A0A336MSL6 A0A0C9RQQ0 A0A3L8E0K2 A0A026WE63 A0A151WEM6 F4X371 A0A158NM94 A0A232FHY5 A0A151IVY5 A0A195FHW7 A0A195BER4 A0A2S2R7B8
PDB
2I13
E-value=7.40105e-41,
Score=422
Ontologies
GO
Topology
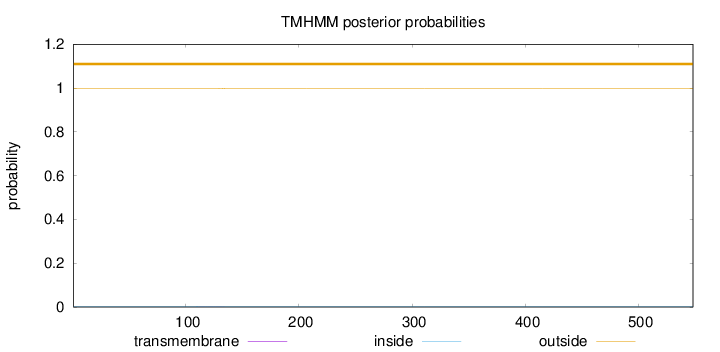
Length:
548
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0181
Exp number, first 60 AAs:
0.00025
Total prob of N-in:
0.00307
outside
1 - 548
Population Genetic Test Statistics
Pi
220.044317
Theta
182.256651
Tajima's D
0.498102
CLR
0.287235
CSRT
0.509324533773311
Interpretation
Uncertain
