Gene
KWMTBOMO08005
Annotation
Zinc_finger_protein_316_[Papilio_machaon]
Transcription factor
Location in the cell
Nuclear Reliability : 3.621
Sequence
CDS
ATGTACCAACACACTGGAATAAAATCTTTTAAATGTAACAAATGTTTCAAAGAATTTATTACGCACTATGAAAAGAGGAAACATATGAGATGTCACAAAATTTATATCTGTGAAGAGTGCAAGAAACAGTTTGATAAATACAATGATTTTCAAAAACATAAAAAGGAACATGATACCAAAGAATATTGCTGTGATAAATGTGGTGTTAGTTTTAAGGAAAGAATACTCATTGTTCGCCATCTGAAAAAACATTCCCAAAGTAATAATCGACATTTTCCTTGCCCTCACAAAAATTGTGGAAGAACTTATTCAAGGAATAGTAATCTAAAACAGCATATTTTAACAAAGCATGAACAAATAACTCATGATTGTCACATATGTAACACTCGATTGTGTTCAAAAGCCAAACTGAATAATCATTTAAAACTACACAATAAACCGATCACGGAAGTAGTTTCTGTACGTGCAAATACCAAAGCGGCTGGTCGCAAAACTCGAAAGGATATAGGCGTTCCGAGGAAGTCTATTGCATTGAAACTAGCCGGCTTTAAAGAACAAAACGAAATAAAATCAGCTATACCATATTTAGCAGAAACGTGTATGACCTAG
Protein
MYQHTGIKSFKCNKCFKEFITHYEKRKHMRCHKIYICEECKKQFDKYNDFQKHKKEHDTKEYCCDKCGVSFKERILIVRHLKKHSQSNNRHFPCPHKNCGRTYSRNSNLKQHILTKHEQITHDCHICNTRLCSKAKLNNHLKLHNKPITEVVSVRANTKAAGRKTRKDIGVPRKSIALKLAGFKEQNEIKSAIPYLAETCMT
Summary
Uniprot
A0A1E1W727
A0A1E1W5P4
A0A0N1INQ6
A0A1E1W1D7
A0A194QC01
A0A212FNZ1
+ More
A0A2A4J8I2 A0A2H1V421 A0A2J7R8T9 A0A026W0N5 F4X1T8 A0A293N6B4 A0A195C6U8 K7JIF6 A0A195BQT8 B4M1N4 A0A158NE70 A0A232EKY1 A0A0M4EYX7 A0A151X4G2 A0A1I8M7V6 A0A195ESD7 A0A1Q3EZY3 A0A0C9R2X4 B0X3D6 A0A0P5X794 H2L3B4 A0A0P5DXE8 A0A0P5CSF6 A0A0P5EBN9 A0A0P6BER7 A0A195D803 A0A0P4Y085 A0A1B0BBG6 A0A1A9XX04 Q1DGG6 E9HBN8 A0A0P6G936 A0A0L0C650 A0A0P6DY10 A0A0P5JZ41 A0A1I7VFB8 A0A0P5RZE7 A0A2A3EP55 Q16HM7 B3MWC0 A0A0G2KCQ2 A0A087ZRK1 A0A1E1XVP2 E2AX63 A0A182HAN9 A0A023G8V5 G3MR60 A0A1E1XCR0 A0A3P9N258 C3ZVG8 A0A3P9KP31 A0A1A9ZII5 A0A310SED6 A0A096MIS1 A0A1Z5L971 A0A3P8SL36 A0A2R5LFR9 A0A3P8SKP7 A0A1B0DA08 A0A1I8PMN1 A0A3Q3EFN0 C3ZSW2 A0A1J4KT44 A0A0P5CKN9 A0A0P5LKK6 A0A0P4ZYG2 A0A2I4BWN0 B4JP09 A0A3N0XT22 B4L2U8 A0A3B3CII4 A0A3Q1JGL8 C3ZDL1 A0A1S0TGT8 D2A389 B4NPP4 A0A3P8T634 A0A146R7X6 A0A1I7VFH7 A0A3Q4IGM4 A0A3B3CJB8 A0A3P8R4S9 C3ZNY3 E9IGU9 A0A287B1N6 A0A3Q7PLX8 A0A1B6GPA5 A0A286ZMW6 A0A3B3VEN3 I3LQ98
A0A2A4J8I2 A0A2H1V421 A0A2J7R8T9 A0A026W0N5 F4X1T8 A0A293N6B4 A0A195C6U8 K7JIF6 A0A195BQT8 B4M1N4 A0A158NE70 A0A232EKY1 A0A0M4EYX7 A0A151X4G2 A0A1I8M7V6 A0A195ESD7 A0A1Q3EZY3 A0A0C9R2X4 B0X3D6 A0A0P5X794 H2L3B4 A0A0P5DXE8 A0A0P5CSF6 A0A0P5EBN9 A0A0P6BER7 A0A195D803 A0A0P4Y085 A0A1B0BBG6 A0A1A9XX04 Q1DGG6 E9HBN8 A0A0P6G936 A0A0L0C650 A0A0P6DY10 A0A0P5JZ41 A0A1I7VFB8 A0A0P5RZE7 A0A2A3EP55 Q16HM7 B3MWC0 A0A0G2KCQ2 A0A087ZRK1 A0A1E1XVP2 E2AX63 A0A182HAN9 A0A023G8V5 G3MR60 A0A1E1XCR0 A0A3P9N258 C3ZVG8 A0A3P9KP31 A0A1A9ZII5 A0A310SED6 A0A096MIS1 A0A1Z5L971 A0A3P8SL36 A0A2R5LFR9 A0A3P8SKP7 A0A1B0DA08 A0A1I8PMN1 A0A3Q3EFN0 C3ZSW2 A0A1J4KT44 A0A0P5CKN9 A0A0P5LKK6 A0A0P4ZYG2 A0A2I4BWN0 B4JP09 A0A3N0XT22 B4L2U8 A0A3B3CII4 A0A3Q1JGL8 C3ZDL1 A0A1S0TGT8 D2A389 B4NPP4 A0A3P8T634 A0A146R7X6 A0A1I7VFH7 A0A3Q4IGM4 A0A3B3CJB8 A0A3P8R4S9 C3ZNY3 E9IGU9 A0A287B1N6 A0A3Q7PLX8 A0A1B6GPA5 A0A286ZMW6 A0A3B3VEN3 I3LQ98
Pubmed
EMBL
GDQN01008281
JAT82773.1
GDQN01008863
JAT82191.1
KQ460937
KPJ10698.1
+ More
GDQN01010247 JAT80807.1 KQ459439 KPJ00956.1 AGBW02004200 OWR55461.1 NWSH01002453 PCG68271.1 ODYU01000577 SOQ35590.1 NEVH01006721 PNF37253.1 KK107503 EZA49625.1 GL888551 EGI59595.1 GFWV01023232 MAA47959.1 KQ978251 KYM95911.1 KQ976424 KYM88503.1 CH940651 EDW65588.1 ADTU01013079 ADTU01013080 NNAY01003710 OXU18972.1 CP012528 ALC49255.1 KQ982548 KYQ55306.1 KQ981993 KYN30817.1 GFDL01014196 JAV20849.1 GBZX01001375 JAG91365.1 DS232312 EDS39828.1 GDIP01076962 JAM26753.1 GDIP01151107 JAJ72295.1 GDIP01170244 JAJ53158.1 GDIP01143855 LRGB01000084 JAJ79547.1 KZS21108.1 GDIP01016179 JAM87536.1 KQ981153 KYN09001.1 GDIP01243909 JAI79492.1 JXJN01011439 CH901889 EAT32245.1 GL732616 EFX70877.1 GDIQ01036880 JAN57857.1 JRES01000948 KNC26879.1 GDIQ01071058 JAN23679.1 GDIQ01192060 JAK59665.1 GDIQ01101161 JAL50565.1 KZ288202 PBC33480.1 CH478159 EAT33755.1 CH902625 EDV35265.1 CU467646 GFAA01000275 JAU03160.1 GL443520 EFN61946.1 JXUM01123026 KQ566670 KXJ69922.1 GBBM01005176 JAC30242.1 JO844361 AEO35978.1 GFAC01002153 JAT97035.1 GG666689 EEN43489.1 KQ770716 OAD52599.1 AYCK01011382 GFJQ02003232 JAW03738.1 GGLE01004226 MBY08352.1 AJVK01028707 GG666675 EEN44307.1 MLAK01000363 OHT14463.1 GDIP01170243 JAJ53159.1 GDIQ01193168 JAK58557.1 GDIP01210692 JAJ12710.1 CH916371 EDV92452.1 RJVU01062164 ROJ30481.1 CH933810 EDW06915.1 GG666612 EEN48817.1 JH712721 EFO13584.1 KQ971338 EFA02279.2 CH964291 EDW86484.1 GCES01122232 JAQ64090.1 GG666654 EEN45718.1 GL763054 EFZ20270.1 AEMK02000012 GECZ01005518 JAS64251.1 DQIR01118914 DQIR01120388 HDA74390.1
GDQN01010247 JAT80807.1 KQ459439 KPJ00956.1 AGBW02004200 OWR55461.1 NWSH01002453 PCG68271.1 ODYU01000577 SOQ35590.1 NEVH01006721 PNF37253.1 KK107503 EZA49625.1 GL888551 EGI59595.1 GFWV01023232 MAA47959.1 KQ978251 KYM95911.1 KQ976424 KYM88503.1 CH940651 EDW65588.1 ADTU01013079 ADTU01013080 NNAY01003710 OXU18972.1 CP012528 ALC49255.1 KQ982548 KYQ55306.1 KQ981993 KYN30817.1 GFDL01014196 JAV20849.1 GBZX01001375 JAG91365.1 DS232312 EDS39828.1 GDIP01076962 JAM26753.1 GDIP01151107 JAJ72295.1 GDIP01170244 JAJ53158.1 GDIP01143855 LRGB01000084 JAJ79547.1 KZS21108.1 GDIP01016179 JAM87536.1 KQ981153 KYN09001.1 GDIP01243909 JAI79492.1 JXJN01011439 CH901889 EAT32245.1 GL732616 EFX70877.1 GDIQ01036880 JAN57857.1 JRES01000948 KNC26879.1 GDIQ01071058 JAN23679.1 GDIQ01192060 JAK59665.1 GDIQ01101161 JAL50565.1 KZ288202 PBC33480.1 CH478159 EAT33755.1 CH902625 EDV35265.1 CU467646 GFAA01000275 JAU03160.1 GL443520 EFN61946.1 JXUM01123026 KQ566670 KXJ69922.1 GBBM01005176 JAC30242.1 JO844361 AEO35978.1 GFAC01002153 JAT97035.1 GG666689 EEN43489.1 KQ770716 OAD52599.1 AYCK01011382 GFJQ02003232 JAW03738.1 GGLE01004226 MBY08352.1 AJVK01028707 GG666675 EEN44307.1 MLAK01000363 OHT14463.1 GDIP01170243 JAJ53159.1 GDIQ01193168 JAK58557.1 GDIP01210692 JAJ12710.1 CH916371 EDV92452.1 RJVU01062164 ROJ30481.1 CH933810 EDW06915.1 GG666612 EEN48817.1 JH712721 EFO13584.1 KQ971338 EFA02279.2 CH964291 EDW86484.1 GCES01122232 JAQ64090.1 GG666654 EEN45718.1 GL763054 EFZ20270.1 AEMK02000012 GECZ01005518 JAS64251.1 DQIR01118914 DQIR01120388 HDA74390.1
Proteomes
UP000053240
UP000053268
UP000007151
UP000218220
UP000235965
UP000053097
+ More
UP000007755 UP000078542 UP000002358 UP000078540 UP000008792 UP000005205 UP000215335 UP000092553 UP000075809 UP000095301 UP000078541 UP000002320 UP000001038 UP000076858 UP000078492 UP000092460 UP000092443 UP000008820 UP000000305 UP000037069 UP000095285 UP000242457 UP000007801 UP000000437 UP000005203 UP000000311 UP000069940 UP000249989 UP000242638 UP000001554 UP000265180 UP000092445 UP000028760 UP000265080 UP000092462 UP000095300 UP000264800 UP000192220 UP000001070 UP000009192 UP000261560 UP000265040 UP000007266 UP000007798 UP000261580 UP000265100 UP000008227 UP000286641 UP000261500
UP000007755 UP000078542 UP000002358 UP000078540 UP000008792 UP000005205 UP000215335 UP000092553 UP000075809 UP000095301 UP000078541 UP000002320 UP000001038 UP000076858 UP000078492 UP000092460 UP000092443 UP000008820 UP000000305 UP000037069 UP000095285 UP000242457 UP000007801 UP000000437 UP000005203 UP000000311 UP000069940 UP000249989 UP000242638 UP000001554 UP000265180 UP000092445 UP000028760 UP000265080 UP000092462 UP000095300 UP000264800 UP000192220 UP000001070 UP000009192 UP000261560 UP000265040 UP000007266 UP000007798 UP000261580 UP000265100 UP000008227 UP000286641 UP000261500
PRIDE
Pfam
Interpro
IPR013087
Znf_C2H2_type
+ More
IPR036236 Znf_C2H2_sf
IPR036691 Endo/exonu/phosph_ase_sf
IPR042228 Dynein_2_C
IPR041658 AAA_lid_11
IPR035699 AAA_6
IPR013594 Dynein_heavy_dom-1
IPR004273 Dynein_heavy_D6_P-loop
IPR013602 Dynein_heavy_dom-2
IPR042219 AAA_lid_11_sf
IPR003593 AAA+_ATPase
IPR027417 P-loop_NTPase
IPR024317 Dynein_heavy_chain_D4_dom
IPR024743 Dynein_HC_stalk
IPR035706 AAA_9
IPR042222 Dynein_2_N
IPR041042 Znf_Hakai
IPR041697 Znf-C2H2_11
IPR036236 Znf_C2H2_sf
IPR036691 Endo/exonu/phosph_ase_sf
IPR042228 Dynein_2_C
IPR041658 AAA_lid_11
IPR035699 AAA_6
IPR013594 Dynein_heavy_dom-1
IPR004273 Dynein_heavy_D6_P-loop
IPR013602 Dynein_heavy_dom-2
IPR042219 AAA_lid_11_sf
IPR003593 AAA+_ATPase
IPR027417 P-loop_NTPase
IPR024317 Dynein_heavy_chain_D4_dom
IPR024743 Dynein_HC_stalk
IPR035706 AAA_9
IPR042222 Dynein_2_N
IPR041042 Znf_Hakai
IPR041697 Znf-C2H2_11
Gene 3D
ProteinModelPortal
A0A1E1W727
A0A1E1W5P4
A0A0N1INQ6
A0A1E1W1D7
A0A194QC01
A0A212FNZ1
+ More
A0A2A4J8I2 A0A2H1V421 A0A2J7R8T9 A0A026W0N5 F4X1T8 A0A293N6B4 A0A195C6U8 K7JIF6 A0A195BQT8 B4M1N4 A0A158NE70 A0A232EKY1 A0A0M4EYX7 A0A151X4G2 A0A1I8M7V6 A0A195ESD7 A0A1Q3EZY3 A0A0C9R2X4 B0X3D6 A0A0P5X794 H2L3B4 A0A0P5DXE8 A0A0P5CSF6 A0A0P5EBN9 A0A0P6BER7 A0A195D803 A0A0P4Y085 A0A1B0BBG6 A0A1A9XX04 Q1DGG6 E9HBN8 A0A0P6G936 A0A0L0C650 A0A0P6DY10 A0A0P5JZ41 A0A1I7VFB8 A0A0P5RZE7 A0A2A3EP55 Q16HM7 B3MWC0 A0A0G2KCQ2 A0A087ZRK1 A0A1E1XVP2 E2AX63 A0A182HAN9 A0A023G8V5 G3MR60 A0A1E1XCR0 A0A3P9N258 C3ZVG8 A0A3P9KP31 A0A1A9ZII5 A0A310SED6 A0A096MIS1 A0A1Z5L971 A0A3P8SL36 A0A2R5LFR9 A0A3P8SKP7 A0A1B0DA08 A0A1I8PMN1 A0A3Q3EFN0 C3ZSW2 A0A1J4KT44 A0A0P5CKN9 A0A0P5LKK6 A0A0P4ZYG2 A0A2I4BWN0 B4JP09 A0A3N0XT22 B4L2U8 A0A3B3CII4 A0A3Q1JGL8 C3ZDL1 A0A1S0TGT8 D2A389 B4NPP4 A0A3P8T634 A0A146R7X6 A0A1I7VFH7 A0A3Q4IGM4 A0A3B3CJB8 A0A3P8R4S9 C3ZNY3 E9IGU9 A0A287B1N6 A0A3Q7PLX8 A0A1B6GPA5 A0A286ZMW6 A0A3B3VEN3 I3LQ98
A0A2A4J8I2 A0A2H1V421 A0A2J7R8T9 A0A026W0N5 F4X1T8 A0A293N6B4 A0A195C6U8 K7JIF6 A0A195BQT8 B4M1N4 A0A158NE70 A0A232EKY1 A0A0M4EYX7 A0A151X4G2 A0A1I8M7V6 A0A195ESD7 A0A1Q3EZY3 A0A0C9R2X4 B0X3D6 A0A0P5X794 H2L3B4 A0A0P5DXE8 A0A0P5CSF6 A0A0P5EBN9 A0A0P6BER7 A0A195D803 A0A0P4Y085 A0A1B0BBG6 A0A1A9XX04 Q1DGG6 E9HBN8 A0A0P6G936 A0A0L0C650 A0A0P6DY10 A0A0P5JZ41 A0A1I7VFB8 A0A0P5RZE7 A0A2A3EP55 Q16HM7 B3MWC0 A0A0G2KCQ2 A0A087ZRK1 A0A1E1XVP2 E2AX63 A0A182HAN9 A0A023G8V5 G3MR60 A0A1E1XCR0 A0A3P9N258 C3ZVG8 A0A3P9KP31 A0A1A9ZII5 A0A310SED6 A0A096MIS1 A0A1Z5L971 A0A3P8SL36 A0A2R5LFR9 A0A3P8SKP7 A0A1B0DA08 A0A1I8PMN1 A0A3Q3EFN0 C3ZSW2 A0A1J4KT44 A0A0P5CKN9 A0A0P5LKK6 A0A0P4ZYG2 A0A2I4BWN0 B4JP09 A0A3N0XT22 B4L2U8 A0A3B3CII4 A0A3Q1JGL8 C3ZDL1 A0A1S0TGT8 D2A389 B4NPP4 A0A3P8T634 A0A146R7X6 A0A1I7VFH7 A0A3Q4IGM4 A0A3B3CJB8 A0A3P8R4S9 C3ZNY3 E9IGU9 A0A287B1N6 A0A3Q7PLX8 A0A1B6GPA5 A0A286ZMW6 A0A3B3VEN3 I3LQ98
PDB
2I13
E-value=5.12728e-11,
Score=159
Ontologies
GO
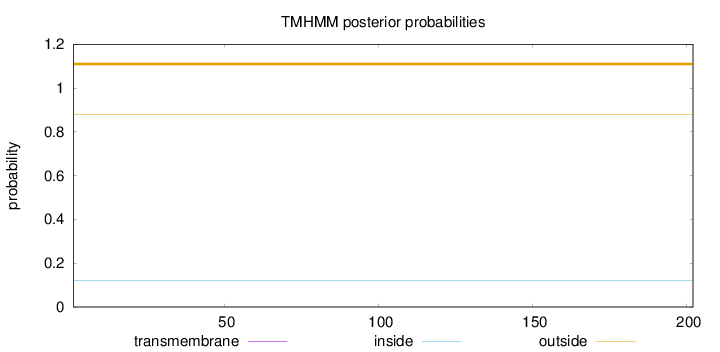
Topology
Length:
202
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.12217
outside
1 - 202
Population Genetic Test Statistics
Pi
218.537591
Theta
178.180225
Tajima's D
0.599974
CLR
0.003992
CSRT
0.537873106344683
Interpretation
Uncertain
