Gene
KWMTBOMO07825
Pre Gene Modal
BGIBMGA000930
Annotation
PREDICTED:_chromatin-remodeling_complex_ATPase_chain_Iswi_isoform_X1_[Bombyx_mori]
Full name
Chromatin-remodeling complex ATPase chain Iswi
Alternative Name
CHRAC 140 kDa subunit
Nucleosome-remodeling factor 140 kDa subunit
Protein imitation swi
Nucleosome-remodeling factor 140 kDa subunit
Protein imitation swi
Transcription factor
Location in the cell
Nuclear Reliability : 3.963
Sequence
CDS
ATGGCCCAGTCGGATGATGCCATGGACGTTGCCGATATTGGCGACAATTCCAATGGCTCTTCAAGTGACACGACATCTTCAAGAGGGAAAGAAGGTGATTTCGAGAGTAAGATTGAAACTGATCGTTCCAAGAGATTCGATTTTCTACTAAAGCAAACGGAGATATTTTCTCATTTTATGACCAACACTCCAAAATCTGGTGGGAGTCCTCCAAAACCAAAGGCTGGCAGACCCAGAAAGATAAAAATAGATACTGAACCTGAAGGTCCTGGAGATCACAGACATAGAAAAACTGAGCAAGAAGAAGATGAAGAACTTCTGGCAGAAACAAATTCAAAACAAAAGACAATATTTAGATTTGAAGCATCTCCACATTACATAAAAAATGGAGAAATGCGGGATTATCAAGTTAGAGGGCTTAACTGGATGATATCATTGTATGAAAATGGTATTAATGGAATTTTGGCTGATGAAATGGGTCTAGGAAAAACTTTGCAAACAATTTCTCTACTTGGATATATGAAACATTTTAAAAACGTTCCTGGTCCTCACATAGTAATTGTACCAAAATCCACTTTGACAAATTGGATGAACGAATTTAAAAAGTGGTGCCCTTCACTAAAAGCTGTGTGTCTTATTGGTGATCAAGAAACAAGGAATACATTTATCCGAGAAACCTTGATGCCTGGAAACTGGGATGTTTGCATTACATCATATGAAATGATTATAAGAGAGAAATCTGTCTTTAAAAAGTTTAACTGGCGTTATATGGTGATTGATGAGGCTCACCGTATCAAGAATGAAAAATCTAAATTATCGGAGCTGCTTAGGGAATTTAAGAGTATGAACAGGTTGCTGTTAACTGGTACTCCCTTGCAAAATAATCTCCATGAGTTGTGGGCACTATTGAATTTCCTTCTGCCTGATGTTTTTAATAGTTCTGATGACTTTGACTCATGGTTTAATACAAATGCAGCCCTAGGTGATAACCAATTGGTTTCACGTTTACATGCTGTACTAAGACCATTTTTGCTTCGGAGACTCAAATCTGAGGTTGAGAAAAAATTAAAGCCTAAAAAGGAATTAAAAGTTTATGTGGGTTTGAGTAAAATGCAAAGAGAGTGGTATACTAAGGTTCTTATGAAGGATATTGATGTAGTAAATGGTGCTGGAAAGGTTGAAAAAATGCGACTCCAGAACATTCTTATGCAGCTACGCAAATGTTGTAACCATCCTTATTTATTTGATGGCGCTGAGCCGGGTCCACCGTACACAACTGATGAACATTTAGTATATAACTGTGGTAAACTTACTATACTGGATAAGCTTTTACCTAAGCTTCAGGAGCAAGAGTCGAGAGTACTTATTTTCTCACAAATGACTAGGATGCTGGACATCTTAGAAGATTATTGCCTTTGGAGGCAATATAAGTATTGCCGTCTAGATGGTCAAACACCACATGAAGATAGAAATCGACAAATAGAAGAGTATAATATGGAGGGGAGTGAGAAATTTGTTTTTATGTTATCTACACGAGCTGGTGGTCTTGGTATTAACTTAACATCAGCTGATGTTGTTATTATTTATGACTCTGATTGGAATCCTCAAATGGATCTGCAGGCCATGGATAGAGCTCACCGTATTGGTCAGAAAAAGCAGGTCAGAGTGTTCCGTTTGATAACAGACAACACGGTTGAAGAAAAGATTGTAGAACGTGCGGAAGTGAAATTGCGCCTGGACAAACTTGTAATTCAGTCCGGTAGACTGGTCGACACTAAGAACCAGCTGAACAAAGATGAGATGCTCAACATGATCAGACATGGTGCCAACCACGTCTTTGCGTCTAAGGATTCTGAGGTTACTGATGAGGACATCGATACGATTTTAGCTAAAGGAGAGAGCAAGACTGAGGAGTTGAAGCAGAAATTGGAGAGCCTGGGCGAGTCGTCCTTGCGAGCATTTTCAATGGACACTCCTGGCGCGACCACCGACTCAGTTTATCAATTTGAAGGCGAGGATTACAGAGAGAAGCAAAAAGTTATCCCAATGGGTAGTTGGATAGAGCCGCCTAAGCGCGAACGTAAAGCTAATTATGCCGTGGACGCCTACTTCAGAGAGGCTCTACGTGTATCTGAACCGAAGGCGCCAAAGGTACAGGCTCCGCGTCCTCCGAAGCAGCCCATCGTACAGGATTTCCAGTTTTTCCCACCACGACTTTTTGAGTTGCTTGATCAAGAAATATATCATTATCGCAAAACCTTAGGATATAAAGTTCCACGTAATCCAGAATTGGGTCCAGATGCAGCGAAAATTCAGAGAGAGGAACAGAGAAAAATAGATGAGGCAGAAGCGCTTTCTGAAGAAGAAGTCCAAGAAAAAGAACAATTACTAACTCAAGGTTTCACTAATTGGACAAAACGTGATTTTAACCAGTTCATAAAAGCAAATGAAAAATACGGACGAGACGATATCGAGAATATTGCAAAAGATGTTGAAGGAAAAACACCAGAAGAGGTTATGGAGTACTCAGCTGTATTTTGGGAACGTTGCCACGAACTTCAAGATATAGATCGAATTATGGGCCAGATCGAAAGGGGCGAGGCTAAAATTCAACGACGAGCCTCTATTAAGAAAGCGTTGGACGCTAAAATGGCGCGGTACCGAGCACCATTCCACCAACTTAGAATCTCTTATGGAACAAACAAGGGCAAAAATTATGTTGAAGAGGAAGACAGGTTCTTGGTGTGCATGCTGCACAAGTTAGGATTCGATAAAGAAAACGTATACGAGGAACTTCGGGCTGCGGTACATTCCGCGCCACAGTTTCGTTTCGATTGGTTTCTAAAGTCAAGAACAGCCGTCGAATTACAACGTCGTTGCAATACGCTCATTACTTTAATTGAACGAGAGAATCAAGAGCTTGAAGAGAAAGAACGAGCCGAAAAGAAAAAGAAAAGCGGGAACGTCAATCCAAGTACGCCAGGCGGCAACGCGGCTGGCAAAGGCGCTAACGCAGGCAAACGCAAAGCTGACAACACTACTGATAGTGCACAGAAACAGAAGAAAAAGAAAACTAAGTAA
Protein
MAQSDDAMDVADIGDNSNGSSSDTTSSRGKEGDFESKIETDRSKRFDFLLKQTEIFSHFMTNTPKSGGSPPKPKAGRPRKIKIDTEPEGPGDHRHRKTEQEEDEELLAETNSKQKTIFRFEASPHYIKNGEMRDYQVRGLNWMISLYENGINGILADEMGLGKTLQTISLLGYMKHFKNVPGPHIVIVPKSTLTNWMNEFKKWCPSLKAVCLIGDQETRNTFIRETLMPGNWDVCITSYEMIIREKSVFKKFNWRYMVIDEAHRIKNEKSKLSELLREFKSMNRLLLTGTPLQNNLHELWALLNFLLPDVFNSSDDFDSWFNTNAALGDNQLVSRLHAVLRPFLLRRLKSEVEKKLKPKKELKVYVGLSKMQREWYTKVLMKDIDVVNGAGKVEKMRLQNILMQLRKCCNHPYLFDGAEPGPPYTTDEHLVYNCGKLTILDKLLPKLQEQESRVLIFSQMTRMLDILEDYCLWRQYKYCRLDGQTPHEDRNRQIEEYNMEGSEKFVFMLSTRAGGLGINLTSADVVIIYDSDWNPQMDLQAMDRAHRIGQKKQVRVFRLITDNTVEEKIVERAEVKLRLDKLVIQSGRLVDTKNQLNKDEMLNMIRHGANHVFASKDSEVTDEDIDTILAKGESKTEELKQKLESLGESSLRAFSMDTPGATTDSVYQFEGEDYREKQKVIPMGSWIEPPKRERKANYAVDAYFREALRVSEPKAPKVQAPRPPKQPIVQDFQFFPPRLFELLDQEIYHYRKTLGYKVPRNPELGPDAAKIQREEQRKIDEAEALSEEEVQEKEQLLTQGFTNWTKRDFNQFIKANEKYGRDDIENIAKDVEGKTPEEVMEYSAVFWERCHELQDIDRIMGQIERGEAKIQRRASIKKALDAKMARYRAPFHQLRISYGTNKGKNYVEEEDRFLVCMLHKLGFDKENVYEELRAAVHSAPQFRFDWFLKSRTAVELQRRCNTLITLIERENQELEEKERAEKKKKSGNVNPSTPGGNAAGKGANAGKRKADNTTDSAQKQKKKKTK
Summary
Description
Energy-transducing component of the chromatin-remodeling complexes NURF (nucleosome-remodeling factor), ACF (ATP-utilizing chromatin assembly and remodeling factor), and CHRAC (chromatin accessibility complex). NURF catalyzes ATP-dependent nucleosome sliding and facilitates transcription of chromatin. It is required for homeotic gene expression, proper larval blood cell development, normal male X chromosome morphology, ecdysteroid signaling and metamorphosis.
Subunit
Component of the NURF complex composed of Caf1, E(bx), Nurf-38 and Iswi.
Similarity
Belongs to the SNF2/RAD54 helicase family. ISWI subfamily.
Keywords
3D-structure
ATP-binding
Chromatin regulator
Complete proteome
Direct protein sequencing
Helicase
Hydrolase
Nucleotide-binding
Nucleus
Phosphoprotein
Reference proteome
Repeat
Transcription
Transcription regulation
Feature
chain Chromatin-remodeling complex ATPase chain Iswi
Uniprot
H9IUK0
A0A3S2M3L3
A0A2A4J6A3
A0A194PE21
A0A194RHI3
A0A2A3EKC5
+ More
A0A088AD04 E9J0E8 A0A195D2P0 F4WVR7 A0A195AVE5 A0A158NHJ8 A0A026WY91 A0A195E2G8 A0A0J7KIG5 E2B9I7 E1ZUT6 A0A151WGL1 A0A195ETL2 A0A347ZJC0 A0A067QXN5 A0A1B6F8Z9 A0A1L8DX53 A0A1B6L2G5 A0A1Y1N2R0 A0A0T6AVN9 D2A0V0 A0A1B6LR13 N6TRI1 A0A140EFS4 A0A1B6D198 A0A0V0G5M6 A0A1B6DS76 A0A069DXR6 A0A224X652 A0A023F5M6 A0A0L7LDD4 A0A1A9Y4I8 A0A1A9UHG4 A0A1B0FC35 A0A1B6CZS2 U5EY61 A0A1B6DLC0 A0A0L0BM44 A0A0K8V959 A0A140HIP5 W8BJN0 A0A034W4D0 A0A1B0BDV8 Q17E28 A0A2J7PFH7 A0A1Q3FVQ5 A0A146MER1 B4KSQ1 T1PG32 B4LP81 B4P567 A0A182QS89 Q24368 A0A182PM09 B4JVM2 B4HPM3 B3NRX4 B4MPP2 A0A0J9RB86 A0A182K1E3 A0A1W4VCW9 A0A182HDN8 A0A182NPB7 A0A2H8TUH5 A0A336LUD4 A0A1I8PAI2 A0A336L7N6 A0A182V9B1 A0A182UDP2 A0A182KV13 A0A1J1HS29 A0A154PFP8 A0A2S2N6D8 A0A182WXY1 Q7QE14 A0A182I603 A0A182YAD0 A0A084WNI5 A0A182ILL8 J9JUM4 A0A182FPM1 A0A0M4EHL5 A0A2M4ADG5 A0A3B0JKQ6 A0A182RLT6 W5JX29 A0A2M3Z6B9 A0A2M4BBQ4 A0A2S2PX75 Q293F0 B3MCU3 A0A0P5WKE3 E9GJ81 A0A0P4Y4S5
A0A088AD04 E9J0E8 A0A195D2P0 F4WVR7 A0A195AVE5 A0A158NHJ8 A0A026WY91 A0A195E2G8 A0A0J7KIG5 E2B9I7 E1ZUT6 A0A151WGL1 A0A195ETL2 A0A347ZJC0 A0A067QXN5 A0A1B6F8Z9 A0A1L8DX53 A0A1B6L2G5 A0A1Y1N2R0 A0A0T6AVN9 D2A0V0 A0A1B6LR13 N6TRI1 A0A140EFS4 A0A1B6D198 A0A0V0G5M6 A0A1B6DS76 A0A069DXR6 A0A224X652 A0A023F5M6 A0A0L7LDD4 A0A1A9Y4I8 A0A1A9UHG4 A0A1B0FC35 A0A1B6CZS2 U5EY61 A0A1B6DLC0 A0A0L0BM44 A0A0K8V959 A0A140HIP5 W8BJN0 A0A034W4D0 A0A1B0BDV8 Q17E28 A0A2J7PFH7 A0A1Q3FVQ5 A0A146MER1 B4KSQ1 T1PG32 B4LP81 B4P567 A0A182QS89 Q24368 A0A182PM09 B4JVM2 B4HPM3 B3NRX4 B4MPP2 A0A0J9RB86 A0A182K1E3 A0A1W4VCW9 A0A182HDN8 A0A182NPB7 A0A2H8TUH5 A0A336LUD4 A0A1I8PAI2 A0A336L7N6 A0A182V9B1 A0A182UDP2 A0A182KV13 A0A1J1HS29 A0A154PFP8 A0A2S2N6D8 A0A182WXY1 Q7QE14 A0A182I603 A0A182YAD0 A0A084WNI5 A0A182ILL8 J9JUM4 A0A182FPM1 A0A0M4EHL5 A0A2M4ADG5 A0A3B0JKQ6 A0A182RLT6 W5JX29 A0A2M3Z6B9 A0A2M4BBQ4 A0A2S2PX75 Q293F0 B3MCU3 A0A0P5WKE3 E9GJ81 A0A0P4Y4S5
EC Number
3.6.4.-
Pubmed
19121390
26354079
21282665
21719571
21347285
24508170
+ More
30249741 20798317 24845553 28004739 18362917 19820115 23537049 26873291 26334808 25474469 26227816 26108605 24495485 25348373 17510324 26823975 17994087 25315136 17550304 7908117 10731132 12537572 12537569 8521502 8521501 12502740 16264191 18327897 14536084 18057021 22936249 26483478 20966253 12364791 14747013 17210077 25244985 24438588 20920257 23761445 15632085 23185243 21292972
30249741 20798317 24845553 28004739 18362917 19820115 23537049 26873291 26334808 25474469 26227816 26108605 24495485 25348373 17510324 26823975 17994087 25315136 17550304 7908117 10731132 12537572 12537569 8521502 8521501 12502740 16264191 18327897 14536084 18057021 22936249 26483478 20966253 12364791 14747013 17210077 25244985 24438588 20920257 23761445 15632085 23185243 21292972
EMBL
BABH01000943
BABH01000944
RSAL01000047
RVE50528.1
NWSH01002871
PCG67409.1
+ More
KQ459606 KPI91273.1 KQ460205 KPJ16899.1 KZ288220 PBC32177.1 GL767462 EFZ13700.1 KQ976973 KYN06634.1 GL888393 EGI61737.1 KQ976737 KYM75944.1 ADTU01015861 KK107069 QOIP01000002 EZA60706.1 RLU25869.1 KQ979763 KYN19271.1 LBMM01007091 KMQ90049.1 GL446556 EFN87620.1 GL434297 EFN75028.1 KQ983167 KYQ46964.1 KQ981975 KYN31578.1 FX985688 BBA84426.1 KK853132 KDR10882.1 GECZ01027479 GECZ01023119 GECZ01012067 GECZ01006744 JAS42290.1 JAS46650.1 JAS57702.1 JAS63025.1 GFDF01003088 JAV10996.1 GEBQ01022131 JAT17846.1 GEZM01017029 JAV90955.1 LJIG01022699 KRT79177.1 KQ971338 EFA01629.1 GEBQ01013948 JAT26029.1 APGK01022501 KB740427 ENN80673.1 KT369803 AML25532.1 GEDC01017819 JAS19479.1 GECL01002730 JAP03394.1 GEDC01008780 JAS28518.1 GBGD01000268 JAC88621.1 GFTR01008471 JAW07955.1 GBBI01002242 JAC16470.1 JTDY01001600 KOB73409.1 CCAG010005008 GEDC01018356 JAS18942.1 GANO01000571 JAB59300.1 GEDC01010815 JAS26483.1 JRES01001654 KNC21121.1 GDHF01016922 JAI35392.1 KT364640 AMO13047.1 GAMC01005065 JAC01491.1 GAKP01010339 GAKP01010336 GAKP01010334 GAKP01010332 JAC48618.1 JXJN01012713 CH477287 EAT44710.1 NEVH01025657 PNF15090.1 GFDL01003354 JAV31691.1 GDHC01000351 JAQ18278.1 CH933808 EDW10550.1 KA646843 AFP61472.1 CH940648 EDW60190.1 CM000158 EDW90729.1 KRJ99412.1 AXCN02001819 L27127 AE013599 AY094908 BT044562 CH916375 EDV98490.1 CH480816 EDW47607.1 CH954179 EDV56276.1 KQS62809.1 CH963849 EDW74081.1 CM002911 KMY93291.1 KMY93292.1 JXUM01034587 JXUM01034588 JXUM01034589 KQ561029 KXJ79969.1 GFXV01005656 MBW17461.1 UFQS01000115 UFQT01000115 SSW99899.1 SSX20279.1 UFQS01001832 UFQT01001832 SSX12485.1 SSX31928.1 CVRI01000020 CRK90821.1 KQ434889 KZC10274.1 GGMR01000096 MBY12715.1 AAAB01008848 EAA07020.3 APCN01003539 ATLV01024612 KE525353 KFB51779.1 ABLF02027798 CP012524 ALC42097.1 GGFK01005498 MBW38819.1 OUUW01000001 SPP75940.1 ADMH02000117 ETN67784.1 GGFM01003293 MBW24044.1 GGFJ01001322 MBW50463.1 GGMS01000826 MBY70029.1 CM000071 EAL24661.2 KRT01441.1 KRT01442.1 CH902619 EDV37345.1 KPU76736.1 GDIP01097883 JAM05832.1 GL732547 EFX80401.1 GDIP01232760 GDIP01183470 GDIP01069741 LRGB01000084 JAI90641.1 KZS21056.1
KQ459606 KPI91273.1 KQ460205 KPJ16899.1 KZ288220 PBC32177.1 GL767462 EFZ13700.1 KQ976973 KYN06634.1 GL888393 EGI61737.1 KQ976737 KYM75944.1 ADTU01015861 KK107069 QOIP01000002 EZA60706.1 RLU25869.1 KQ979763 KYN19271.1 LBMM01007091 KMQ90049.1 GL446556 EFN87620.1 GL434297 EFN75028.1 KQ983167 KYQ46964.1 KQ981975 KYN31578.1 FX985688 BBA84426.1 KK853132 KDR10882.1 GECZ01027479 GECZ01023119 GECZ01012067 GECZ01006744 JAS42290.1 JAS46650.1 JAS57702.1 JAS63025.1 GFDF01003088 JAV10996.1 GEBQ01022131 JAT17846.1 GEZM01017029 JAV90955.1 LJIG01022699 KRT79177.1 KQ971338 EFA01629.1 GEBQ01013948 JAT26029.1 APGK01022501 KB740427 ENN80673.1 KT369803 AML25532.1 GEDC01017819 JAS19479.1 GECL01002730 JAP03394.1 GEDC01008780 JAS28518.1 GBGD01000268 JAC88621.1 GFTR01008471 JAW07955.1 GBBI01002242 JAC16470.1 JTDY01001600 KOB73409.1 CCAG010005008 GEDC01018356 JAS18942.1 GANO01000571 JAB59300.1 GEDC01010815 JAS26483.1 JRES01001654 KNC21121.1 GDHF01016922 JAI35392.1 KT364640 AMO13047.1 GAMC01005065 JAC01491.1 GAKP01010339 GAKP01010336 GAKP01010334 GAKP01010332 JAC48618.1 JXJN01012713 CH477287 EAT44710.1 NEVH01025657 PNF15090.1 GFDL01003354 JAV31691.1 GDHC01000351 JAQ18278.1 CH933808 EDW10550.1 KA646843 AFP61472.1 CH940648 EDW60190.1 CM000158 EDW90729.1 KRJ99412.1 AXCN02001819 L27127 AE013599 AY094908 BT044562 CH916375 EDV98490.1 CH480816 EDW47607.1 CH954179 EDV56276.1 KQS62809.1 CH963849 EDW74081.1 CM002911 KMY93291.1 KMY93292.1 JXUM01034587 JXUM01034588 JXUM01034589 KQ561029 KXJ79969.1 GFXV01005656 MBW17461.1 UFQS01000115 UFQT01000115 SSW99899.1 SSX20279.1 UFQS01001832 UFQT01001832 SSX12485.1 SSX31928.1 CVRI01000020 CRK90821.1 KQ434889 KZC10274.1 GGMR01000096 MBY12715.1 AAAB01008848 EAA07020.3 APCN01003539 ATLV01024612 KE525353 KFB51779.1 ABLF02027798 CP012524 ALC42097.1 GGFK01005498 MBW38819.1 OUUW01000001 SPP75940.1 ADMH02000117 ETN67784.1 GGFM01003293 MBW24044.1 GGFJ01001322 MBW50463.1 GGMS01000826 MBY70029.1 CM000071 EAL24661.2 KRT01441.1 KRT01442.1 CH902619 EDV37345.1 KPU76736.1 GDIP01097883 JAM05832.1 GL732547 EFX80401.1 GDIP01232760 GDIP01183470 GDIP01069741 LRGB01000084 JAI90641.1 KZS21056.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000053240
UP000242457
+ More
UP000005203 UP000078542 UP000007755 UP000078540 UP000005205 UP000053097 UP000279307 UP000078492 UP000036403 UP000008237 UP000000311 UP000075809 UP000078541 UP000027135 UP000007266 UP000019118 UP000037510 UP000092443 UP000078200 UP000092444 UP000037069 UP000092460 UP000008820 UP000235965 UP000009192 UP000095301 UP000008792 UP000002282 UP000075886 UP000000803 UP000075885 UP000001070 UP000001292 UP000008711 UP000007798 UP000075881 UP000192221 UP000069940 UP000249989 UP000075884 UP000095300 UP000075903 UP000075902 UP000075882 UP000183832 UP000076502 UP000076407 UP000007062 UP000075840 UP000076408 UP000030765 UP000075880 UP000007819 UP000069272 UP000092553 UP000268350 UP000075900 UP000000673 UP000001819 UP000007801 UP000000305 UP000076858
UP000005203 UP000078542 UP000007755 UP000078540 UP000005205 UP000053097 UP000279307 UP000078492 UP000036403 UP000008237 UP000000311 UP000075809 UP000078541 UP000027135 UP000007266 UP000019118 UP000037510 UP000092443 UP000078200 UP000092444 UP000037069 UP000092460 UP000008820 UP000235965 UP000009192 UP000095301 UP000008792 UP000002282 UP000075886 UP000000803 UP000075885 UP000001070 UP000001292 UP000008711 UP000007798 UP000075881 UP000192221 UP000069940 UP000249989 UP000075884 UP000095300 UP000075903 UP000075902 UP000075882 UP000183832 UP000076502 UP000076407 UP000007062 UP000075840 UP000076408 UP000030765 UP000075880 UP000007819 UP000069272 UP000092553 UP000268350 UP000075900 UP000000673 UP000001819 UP000007801 UP000000305 UP000076858
Pfam
Interpro
IPR029915
ISWI_metazoa
+ More
IPR015195 SLIDE
IPR038718 SNF2-like_sf
IPR027417 P-loop_NTPase
IPR036306 ISWI_HAND-dom_sf
IPR000330 SNF2_N
IPR017884 SANT_dom
IPR001650 Helicase_C
IPR009057 Homeobox-like_sf
IPR015194 ISWI_HAND-dom
IPR001005 SANT/Myb
IPR020838 DBINO
IPR014001 Helicase_ATP-bd
IPR001254 Trypsin_dom
IPR009003 Peptidase_S1_PA
IPR015195 SLIDE
IPR038718 SNF2-like_sf
IPR027417 P-loop_NTPase
IPR036306 ISWI_HAND-dom_sf
IPR000330 SNF2_N
IPR017884 SANT_dom
IPR001650 Helicase_C
IPR009057 Homeobox-like_sf
IPR015194 ISWI_HAND-dom
IPR001005 SANT/Myb
IPR020838 DBINO
IPR014001 Helicase_ATP-bd
IPR001254 Trypsin_dom
IPR009003 Peptidase_S1_PA
Gene 3D
ProteinModelPortal
H9IUK0
A0A3S2M3L3
A0A2A4J6A3
A0A194PE21
A0A194RHI3
A0A2A3EKC5
+ More
A0A088AD04 E9J0E8 A0A195D2P0 F4WVR7 A0A195AVE5 A0A158NHJ8 A0A026WY91 A0A195E2G8 A0A0J7KIG5 E2B9I7 E1ZUT6 A0A151WGL1 A0A195ETL2 A0A347ZJC0 A0A067QXN5 A0A1B6F8Z9 A0A1L8DX53 A0A1B6L2G5 A0A1Y1N2R0 A0A0T6AVN9 D2A0V0 A0A1B6LR13 N6TRI1 A0A140EFS4 A0A1B6D198 A0A0V0G5M6 A0A1B6DS76 A0A069DXR6 A0A224X652 A0A023F5M6 A0A0L7LDD4 A0A1A9Y4I8 A0A1A9UHG4 A0A1B0FC35 A0A1B6CZS2 U5EY61 A0A1B6DLC0 A0A0L0BM44 A0A0K8V959 A0A140HIP5 W8BJN0 A0A034W4D0 A0A1B0BDV8 Q17E28 A0A2J7PFH7 A0A1Q3FVQ5 A0A146MER1 B4KSQ1 T1PG32 B4LP81 B4P567 A0A182QS89 Q24368 A0A182PM09 B4JVM2 B4HPM3 B3NRX4 B4MPP2 A0A0J9RB86 A0A182K1E3 A0A1W4VCW9 A0A182HDN8 A0A182NPB7 A0A2H8TUH5 A0A336LUD4 A0A1I8PAI2 A0A336L7N6 A0A182V9B1 A0A182UDP2 A0A182KV13 A0A1J1HS29 A0A154PFP8 A0A2S2N6D8 A0A182WXY1 Q7QE14 A0A182I603 A0A182YAD0 A0A084WNI5 A0A182ILL8 J9JUM4 A0A182FPM1 A0A0M4EHL5 A0A2M4ADG5 A0A3B0JKQ6 A0A182RLT6 W5JX29 A0A2M3Z6B9 A0A2M4BBQ4 A0A2S2PX75 Q293F0 B3MCU3 A0A0P5WKE3 E9GJ81 A0A0P4Y4S5
A0A088AD04 E9J0E8 A0A195D2P0 F4WVR7 A0A195AVE5 A0A158NHJ8 A0A026WY91 A0A195E2G8 A0A0J7KIG5 E2B9I7 E1ZUT6 A0A151WGL1 A0A195ETL2 A0A347ZJC0 A0A067QXN5 A0A1B6F8Z9 A0A1L8DX53 A0A1B6L2G5 A0A1Y1N2R0 A0A0T6AVN9 D2A0V0 A0A1B6LR13 N6TRI1 A0A140EFS4 A0A1B6D198 A0A0V0G5M6 A0A1B6DS76 A0A069DXR6 A0A224X652 A0A023F5M6 A0A0L7LDD4 A0A1A9Y4I8 A0A1A9UHG4 A0A1B0FC35 A0A1B6CZS2 U5EY61 A0A1B6DLC0 A0A0L0BM44 A0A0K8V959 A0A140HIP5 W8BJN0 A0A034W4D0 A0A1B0BDV8 Q17E28 A0A2J7PFH7 A0A1Q3FVQ5 A0A146MER1 B4KSQ1 T1PG32 B4LP81 B4P567 A0A182QS89 Q24368 A0A182PM09 B4JVM2 B4HPM3 B3NRX4 B4MPP2 A0A0J9RB86 A0A182K1E3 A0A1W4VCW9 A0A182HDN8 A0A182NPB7 A0A2H8TUH5 A0A336LUD4 A0A1I8PAI2 A0A336L7N6 A0A182V9B1 A0A182UDP2 A0A182KV13 A0A1J1HS29 A0A154PFP8 A0A2S2N6D8 A0A182WXY1 Q7QE14 A0A182I603 A0A182YAD0 A0A084WNI5 A0A182ILL8 J9JUM4 A0A182FPM1 A0A0M4EHL5 A0A2M4ADG5 A0A3B0JKQ6 A0A182RLT6 W5JX29 A0A2M3Z6B9 A0A2M4BBQ4 A0A2S2PX75 Q293F0 B3MCU3 A0A0P5WKE3 E9GJ81 A0A0P4Y4S5
PDB
6K1P
E-value=0,
Score=2052
Ontologies
GO
GO:0005524
GO:0003677
GO:0031491
GO:0016887
GO:0043044
GO:0016589
GO:0005634
GO:0004386
GO:0008094
GO:0070615
GO:0016584
GO:0008134
GO:0045944
GO:0008623
GO:0035092
GO:0045892
GO:0031213
GO:0035063
GO:0042766
GO:0035041
GO:0042752
GO:0005667
GO:0006334
GO:0016590
GO:0006351
GO:0035076
GO:0005700
GO:0003678
GO:0006325
GO:0031010
GO:0045893
GO:0006355
GO:0000166
GO:0006333
GO:0006357
GO:0006338
GO:0007283
GO:0004252
GO:0003676
GO:0016818
GO:0003707
GO:0051537
PANTHER
Topology
Subcellular location
Nucleus
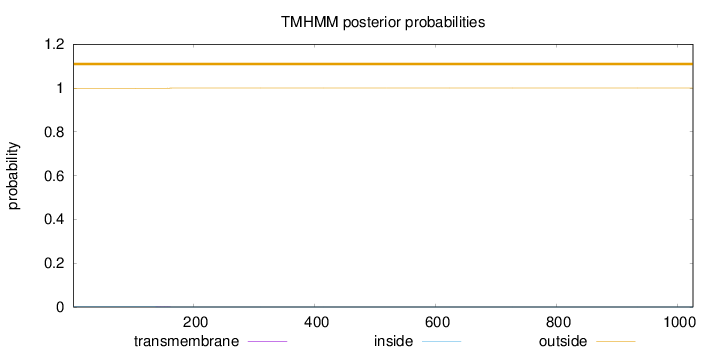
Length:
1026
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02241
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00110
outside
1 - 1026
Population Genetic Test Statistics
Pi
183.192187
Theta
175.589347
Tajima's D
-0.539453
CLR
10.16452
CSRT
0.228438578071096
Interpretation
Uncertain
