Gene
KWMTBOMO06921
Annotation
PREDICTED:_uncharacterized_protein_LOC101739964_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 4.004
Sequence
CDS
ATGGACGAACACTTAGGGGCACCTTCCGGCGCTCGGGAGGCGGCTATTAATGCGTTAGAGCAACACCAGCAGAGCGGCGAGGACACCAGCGACTCTCAGGCCGAGCCGGCGACCCGGGGCAGTCCAAAAAGTTGGACCCCGACTGACATGGACAATGCCTTGGAGGCGCTCAGGAGCCACCACATGAGTTTGACTAAGGCCTCGGCGCTGTTCGGCATCCCGTCCACCACGCTGTGGCAGCGCGCGCACCGCCTCGGCATCGACACGCCCAAGAAGGAGGGCTCCTCGAAGTCTTGGAGCGAGACGGACCTCAAGAACGCGCTGGAAGCTCTCAGGGCCGGCTCTATATCCGCTAACAAGGCCAGCAAGGCTTACGGAATTCCGAGTAGCACTCTGTACAAGATAGCTCGTCGGGAGGGCATCCGTCTCGCGGCGCCCTTCAACGCGGCCCCCACGGCGTGGCGCCGCACCGACCTGCGCCACGCGCTCGGCGCGCTGCGAGCCCGCGCGCTCTCCGTGCAGCGCGCCGCCGCACTGTACGGCATACCCACCGGTACTGTACTCCTCGCACACCGTACTGTACACTCGCACTCCGTACTGTACACCTCGCACTCCGTACTGTACACCTCGCACTCCGTACTGTACACCTCGCACACTGTGTTATACACATGCTGCTTCGAGCTGAGCCTCTATGGCCGCTCAGGTACATTATACGGCCGGTGTAAGCGAGAAGGCATCGAGCTGTCTCGCTCTAACCCGACCCCGTGGTCCGAAGATGCAATGGCGGAAGCACTTGAGGCAGTGAGGGTTGGCCAGATGTCGATAAACCAGGCGGCTATACATTACAACTTGCCGTACTCGTCTCTGTATGGACGGTTCAAAAGATGCAAATACCAGAGCGTGCAACAGTACCAGCAGCCGCCGATTGCCCCGGACGGCCCCCCGCACGACCCGGGGCCCCCCCGGCCGGCCCCCCACCCCCCCGCCTACCCGCACTACCCGCACGCGCACGAGCAGGACTACGCGCAGACGCTGTGCTACCACCATCACTACAATACTGTCATTAGTTGA
Protein
MDEHLGAPSGAREAAINALEQHQQSGEDTSDSQAEPATRGSPKSWTPTDMDNALEALRSHHMSLTKASALFGIPSTTLWQRAHRLGIDTPKKEGSSKSWSETDLKNALEALRAGSISANKASKAYGIPSSTLYKIARREGIRLAAPFNAAPTAWRRTDLRHALGALRARALSVQRAAALYGIPTGTVLLAHRTVHSHSVLYTSHSVLYTSHSVLYTSHTVLYTCCFELSLYGRSGTLYGRCKREGIELSRSNPTPWSEDAMAEALEAVRVGQMSINQAAIHYNLPYSSLYGRFKRCKYQSVQQYQQPPIAPDGPPHDPGPPRPAPHPPAYPHYPHAHEQDYAQTLCYHHHYNTVIS
Summary
Uniprot
A0A3S2NME2
A0A212FLU5
A0A2H1V3C9
A0A2A4K7N0
A0A1B6CFC2
A0A1B6D8Y0
+ More
A0A2J7R9R1 A0A0C9RFL0 A0A2J7R9Q3 A0A0C9RA61 A0A1J1HPH9 A0A182T527 A0A1B6C9Y4 A0A2P8XU84 T1E1G8 A0A1B6I1P3 N6T0G0 A0A1B6C4I0 U4UAX6 A0A182TVJ8 A0A1Y1MP10 A0A1Q3EU61 A0A084WRW5 A0A2J7R9R0 A0A1Q3EU96 A0A2J7R9Q1 A0A182LGQ7 A0A224XM60 A0A182QU63 A0A182V9A9 A0A1Y1ML28 A0A1Y1ML43 B0X4F4 V9IHK9 A0A0C9RJ77 E2AZB9 D6WFD6 F4WH63 A0A232EVR6 A0A0C9RZ25 A0A195CRW5 E9INS0 A0A0L0BR42 U4U8T6 A0A336MTT4 A0A1W4X9Y3 A0A336KXX4 E0VAP3 A0A158NTB2 A0A1B6K1R6 A0A1A9XNG4 A0A182HNV0 A0A1B0B098 A0A1W4XJ71 A0A1W4XK51 A0A0J7KLF0 A0A023F2H9 A0A1A9WHD7 A0A3L8D3I1 A0A0Q9X7J1 A0A0M9A070 A0A1Y1ML35 A0A1Y1ML56 W8BIA3 Q0E9C8 A0A1B6KZB8 Q7JP00 A0A034VRT1 A0A195EVX6 A0A151WS56 Q24457 K7IP00 A0A034VMG9 A4UZC9 A0A0P4VVS1 A0A0A1XFI0 A0A088ANV3 T1GE77 A0A182FCU9 A0A1A9VFG3 A0A154PQZ6 E2BJ90 A0A182MKL3 A0A1S4G5K3 A0A1B0AHN3 J9HIT3 A0NDV3 A0A0A1X5I5 T1I9J7 A0A1B0FIY3 A0A2S2NMR7 A0A182WMF1 A0A182KFZ3 A0A182NQ87 A0A2A3EQW6 J9HZ80 O77168 A0A195BUL5 A0A0L7RB50 A0A0K8V3V4
A0A2J7R9R1 A0A0C9RFL0 A0A2J7R9Q3 A0A0C9RA61 A0A1J1HPH9 A0A182T527 A0A1B6C9Y4 A0A2P8XU84 T1E1G8 A0A1B6I1P3 N6T0G0 A0A1B6C4I0 U4UAX6 A0A182TVJ8 A0A1Y1MP10 A0A1Q3EU61 A0A084WRW5 A0A2J7R9R0 A0A1Q3EU96 A0A2J7R9Q1 A0A182LGQ7 A0A224XM60 A0A182QU63 A0A182V9A9 A0A1Y1ML28 A0A1Y1ML43 B0X4F4 V9IHK9 A0A0C9RJ77 E2AZB9 D6WFD6 F4WH63 A0A232EVR6 A0A0C9RZ25 A0A195CRW5 E9INS0 A0A0L0BR42 U4U8T6 A0A336MTT4 A0A1W4X9Y3 A0A336KXX4 E0VAP3 A0A158NTB2 A0A1B6K1R6 A0A1A9XNG4 A0A182HNV0 A0A1B0B098 A0A1W4XJ71 A0A1W4XK51 A0A0J7KLF0 A0A023F2H9 A0A1A9WHD7 A0A3L8D3I1 A0A0Q9X7J1 A0A0M9A070 A0A1Y1ML35 A0A1Y1ML56 W8BIA3 Q0E9C8 A0A1B6KZB8 Q7JP00 A0A034VRT1 A0A195EVX6 A0A151WS56 Q24457 K7IP00 A0A034VMG9 A4UZC9 A0A0P4VVS1 A0A0A1XFI0 A0A088ANV3 T1GE77 A0A182FCU9 A0A1A9VFG3 A0A154PQZ6 E2BJ90 A0A182MKL3 A0A1S4G5K3 A0A1B0AHN3 J9HIT3 A0NDV3 A0A0A1X5I5 T1I9J7 A0A1B0FIY3 A0A2S2NMR7 A0A182WMF1 A0A182KFZ3 A0A182NQ87 A0A2A3EQW6 J9HZ80 O77168 A0A195BUL5 A0A0L7RB50 A0A0K8V3V4
Pubmed
22118469
29403074
24330624
23537049
28004739
24438588
+ More
20966253 20798317 18362917 19820115 21719571 28648823 21282665 26108605 20566863 21347285 25474469 30249741 17994087 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 8674425 25348373 8557044 20075255 27129103 25830018 17510324 12364791 14747013 17210077 9774480
20966253 20798317 18362917 19820115 21719571 28648823 21282665 26108605 20566863 21347285 25474469 30249741 17994087 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 8674425 25348373 8557044 20075255 27129103 25830018 17510324 12364791 14747013 17210077 9774480
EMBL
RSAL01000034
RVE51379.1
AGBW02007696
OWR54706.1
ODYU01000452
SOQ35276.1
+ More
NWSH01000092 PCG79660.1 GEDC01025243 JAS12055.1 GEDC01015149 JAS22149.1 NEVH01006580 PNF37570.1 GBYB01007175 JAG76942.1 PNF37568.1 GBYB01013279 GBYB01013280 JAG83046.1 JAG83047.1 CVRI01000006 CRK88121.1 GEDC01027001 JAS10297.1 PYGN01001337 PSN35577.1 GALA01001684 JAA93168.1 GECU01026880 JAS80826.1 APGK01049565 KB741156 ENN73544.1 GEDC01029143 JAS08155.1 KB632169 ERL89463.1 GEZM01028166 GEZM01028162 GEZM01028159 JAV86420.1 GFDL01016213 JAV18832.1 ATLV01026257 KE525409 KFB52959.1 PNF37569.1 GFDL01016219 JAV18826.1 PNF37567.1 GFTR01006744 JAW09682.1 AXCN02001015 GEZM01028165 GEZM01028164 JAV86411.1 GEZM01028161 GEZM01028158 JAV86422.1 DS232338 EDS40307.1 JR047949 AEY60545.1 GBYB01013282 JAG83049.1 GL444196 EFN61217.1 KQ971319 EFA00489.2 GL888148 EGI66542.1 NNAY01001962 OXU22436.1 GBYB01013281 JAG83048.1 KQ977329 KYN03483.1 GL764397 EFZ17700.1 JRES01001497 KNC22453.1 ERL89462.1 UFQS01001182 UFQT01001182 SSX09582.1 SSX29378.1 SSX09581.1 SSX29377.1 DS235012 EEB10449.1 ADTU01025671 ADTU01025672 ADTU01025673 ADTU01025674 ADTU01025675 ADTU01025676 ADTU01025677 GECU01002297 JAT05410.1 APCN01002791 JXJN01006613 JXJN01006614 LBMM01005917 KMQ91097.1 GBBI01003202 JAC15510.1 QOIP01000014 RLU14806.1 CH933808 KRG04303.1 KQ435783 KOX74685.1 GEZM01028167 GEZM01028163 JAV86414.1 GEZM01028160 GEZM01028157 JAV86424.1 GAMC01009842 JAB96713.1 AE013599 AAM68769.1 AAM68770.1 AAO41432.1 AAO41433.1 AAO41434.1 GEBQ01023189 JAT16788.1 U48402 AAC47154.1 GAKP01014417 JAC44535.1 KQ981954 KYN32044.1 KQ982794 KYQ50621.1 X90986 CAA62475.1 GAKP01014416 JAC44536.1 AAX52713.1 AAX52714.1 GDKW01000437 JAI56158.1 GBXI01004575 JAD09717.1 CAQQ02045984 CAQQ02045985 CAQQ02045986 CAQQ02045987 CAQQ02045988 CAQQ02045989 CAQQ02045990 CAQQ02045991 KQ435007 KZC13540.1 GL448558 EFN84240.1 AXCM01010231 CH477565 EJY57794.1 AAAB01008900 EAU76837.2 GBXI01008364 JAD05928.1 ACPB03022206 CCAG010022916 GGMR01005864 MBY18483.1 KZ288193 PBC34080.1 EJY57795.1 AF084556 AAC71015.1 KQ976409 KYM90886.1 KQ414618 KOC67966.1 GDHF01019079 GDHF01010960 JAI33235.1 JAI41354.1
NWSH01000092 PCG79660.1 GEDC01025243 JAS12055.1 GEDC01015149 JAS22149.1 NEVH01006580 PNF37570.1 GBYB01007175 JAG76942.1 PNF37568.1 GBYB01013279 GBYB01013280 JAG83046.1 JAG83047.1 CVRI01000006 CRK88121.1 GEDC01027001 JAS10297.1 PYGN01001337 PSN35577.1 GALA01001684 JAA93168.1 GECU01026880 JAS80826.1 APGK01049565 KB741156 ENN73544.1 GEDC01029143 JAS08155.1 KB632169 ERL89463.1 GEZM01028166 GEZM01028162 GEZM01028159 JAV86420.1 GFDL01016213 JAV18832.1 ATLV01026257 KE525409 KFB52959.1 PNF37569.1 GFDL01016219 JAV18826.1 PNF37567.1 GFTR01006744 JAW09682.1 AXCN02001015 GEZM01028165 GEZM01028164 JAV86411.1 GEZM01028161 GEZM01028158 JAV86422.1 DS232338 EDS40307.1 JR047949 AEY60545.1 GBYB01013282 JAG83049.1 GL444196 EFN61217.1 KQ971319 EFA00489.2 GL888148 EGI66542.1 NNAY01001962 OXU22436.1 GBYB01013281 JAG83048.1 KQ977329 KYN03483.1 GL764397 EFZ17700.1 JRES01001497 KNC22453.1 ERL89462.1 UFQS01001182 UFQT01001182 SSX09582.1 SSX29378.1 SSX09581.1 SSX29377.1 DS235012 EEB10449.1 ADTU01025671 ADTU01025672 ADTU01025673 ADTU01025674 ADTU01025675 ADTU01025676 ADTU01025677 GECU01002297 JAT05410.1 APCN01002791 JXJN01006613 JXJN01006614 LBMM01005917 KMQ91097.1 GBBI01003202 JAC15510.1 QOIP01000014 RLU14806.1 CH933808 KRG04303.1 KQ435783 KOX74685.1 GEZM01028167 GEZM01028163 JAV86414.1 GEZM01028160 GEZM01028157 JAV86424.1 GAMC01009842 JAB96713.1 AE013599 AAM68769.1 AAM68770.1 AAO41432.1 AAO41433.1 AAO41434.1 GEBQ01023189 JAT16788.1 U48402 AAC47154.1 GAKP01014417 JAC44535.1 KQ981954 KYN32044.1 KQ982794 KYQ50621.1 X90986 CAA62475.1 GAKP01014416 JAC44536.1 AAX52713.1 AAX52714.1 GDKW01000437 JAI56158.1 GBXI01004575 JAD09717.1 CAQQ02045984 CAQQ02045985 CAQQ02045986 CAQQ02045987 CAQQ02045988 CAQQ02045989 CAQQ02045990 CAQQ02045991 KQ435007 KZC13540.1 GL448558 EFN84240.1 AXCM01010231 CH477565 EJY57794.1 AAAB01008900 EAU76837.2 GBXI01008364 JAD05928.1 ACPB03022206 CCAG010022916 GGMR01005864 MBY18483.1 KZ288193 PBC34080.1 EJY57795.1 AF084556 AAC71015.1 KQ976409 KYM90886.1 KQ414618 KOC67966.1 GDHF01019079 GDHF01010960 JAI33235.1 JAI41354.1
Proteomes
UP000283053
UP000007151
UP000218220
UP000235965
UP000183832
UP000075901
+ More
UP000245037 UP000019118 UP000030742 UP000075902 UP000030765 UP000075882 UP000075886 UP000075903 UP000002320 UP000000311 UP000007266 UP000007755 UP000215335 UP000078542 UP000037069 UP000192223 UP000009046 UP000005205 UP000092443 UP000075840 UP000092460 UP000036403 UP000091820 UP000279307 UP000009192 UP000053105 UP000000803 UP000078541 UP000075809 UP000002358 UP000005203 UP000015102 UP000069272 UP000078200 UP000076502 UP000008237 UP000075883 UP000092445 UP000008820 UP000007062 UP000015103 UP000092444 UP000075920 UP000075881 UP000075884 UP000242457 UP000078540 UP000053825
UP000245037 UP000019118 UP000030742 UP000075902 UP000030765 UP000075882 UP000075886 UP000075903 UP000002320 UP000000311 UP000007266 UP000007755 UP000215335 UP000078542 UP000037069 UP000192223 UP000009046 UP000005205 UP000092443 UP000075840 UP000092460 UP000036403 UP000091820 UP000279307 UP000009192 UP000053105 UP000000803 UP000078541 UP000075809 UP000002358 UP000005203 UP000015102 UP000069272 UP000078200 UP000076502 UP000008237 UP000075883 UP000092445 UP000008820 UP000007062 UP000015103 UP000092444 UP000075920 UP000075881 UP000075884 UP000242457 UP000078540 UP000053825
Interpro
ProteinModelPortal
A0A3S2NME2
A0A212FLU5
A0A2H1V3C9
A0A2A4K7N0
A0A1B6CFC2
A0A1B6D8Y0
+ More
A0A2J7R9R1 A0A0C9RFL0 A0A2J7R9Q3 A0A0C9RA61 A0A1J1HPH9 A0A182T527 A0A1B6C9Y4 A0A2P8XU84 T1E1G8 A0A1B6I1P3 N6T0G0 A0A1B6C4I0 U4UAX6 A0A182TVJ8 A0A1Y1MP10 A0A1Q3EU61 A0A084WRW5 A0A2J7R9R0 A0A1Q3EU96 A0A2J7R9Q1 A0A182LGQ7 A0A224XM60 A0A182QU63 A0A182V9A9 A0A1Y1ML28 A0A1Y1ML43 B0X4F4 V9IHK9 A0A0C9RJ77 E2AZB9 D6WFD6 F4WH63 A0A232EVR6 A0A0C9RZ25 A0A195CRW5 E9INS0 A0A0L0BR42 U4U8T6 A0A336MTT4 A0A1W4X9Y3 A0A336KXX4 E0VAP3 A0A158NTB2 A0A1B6K1R6 A0A1A9XNG4 A0A182HNV0 A0A1B0B098 A0A1W4XJ71 A0A1W4XK51 A0A0J7KLF0 A0A023F2H9 A0A1A9WHD7 A0A3L8D3I1 A0A0Q9X7J1 A0A0M9A070 A0A1Y1ML35 A0A1Y1ML56 W8BIA3 Q0E9C8 A0A1B6KZB8 Q7JP00 A0A034VRT1 A0A195EVX6 A0A151WS56 Q24457 K7IP00 A0A034VMG9 A4UZC9 A0A0P4VVS1 A0A0A1XFI0 A0A088ANV3 T1GE77 A0A182FCU9 A0A1A9VFG3 A0A154PQZ6 E2BJ90 A0A182MKL3 A0A1S4G5K3 A0A1B0AHN3 J9HIT3 A0NDV3 A0A0A1X5I5 T1I9J7 A0A1B0FIY3 A0A2S2NMR7 A0A182WMF1 A0A182KFZ3 A0A182NQ87 A0A2A3EQW6 J9HZ80 O77168 A0A195BUL5 A0A0L7RB50 A0A0K8V3V4
A0A2J7R9R1 A0A0C9RFL0 A0A2J7R9Q3 A0A0C9RA61 A0A1J1HPH9 A0A182T527 A0A1B6C9Y4 A0A2P8XU84 T1E1G8 A0A1B6I1P3 N6T0G0 A0A1B6C4I0 U4UAX6 A0A182TVJ8 A0A1Y1MP10 A0A1Q3EU61 A0A084WRW5 A0A2J7R9R0 A0A1Q3EU96 A0A2J7R9Q1 A0A182LGQ7 A0A224XM60 A0A182QU63 A0A182V9A9 A0A1Y1ML28 A0A1Y1ML43 B0X4F4 V9IHK9 A0A0C9RJ77 E2AZB9 D6WFD6 F4WH63 A0A232EVR6 A0A0C9RZ25 A0A195CRW5 E9INS0 A0A0L0BR42 U4U8T6 A0A336MTT4 A0A1W4X9Y3 A0A336KXX4 E0VAP3 A0A158NTB2 A0A1B6K1R6 A0A1A9XNG4 A0A182HNV0 A0A1B0B098 A0A1W4XJ71 A0A1W4XK51 A0A0J7KLF0 A0A023F2H9 A0A1A9WHD7 A0A3L8D3I1 A0A0Q9X7J1 A0A0M9A070 A0A1Y1ML35 A0A1Y1ML56 W8BIA3 Q0E9C8 A0A1B6KZB8 Q7JP00 A0A034VRT1 A0A195EVX6 A0A151WS56 Q24457 K7IP00 A0A034VMG9 A4UZC9 A0A0P4VVS1 A0A0A1XFI0 A0A088ANV3 T1GE77 A0A182FCU9 A0A1A9VFG3 A0A154PQZ6 E2BJ90 A0A182MKL3 A0A1S4G5K3 A0A1B0AHN3 J9HIT3 A0NDV3 A0A0A1X5I5 T1I9J7 A0A1B0FIY3 A0A2S2NMR7 A0A182WMF1 A0A182KFZ3 A0A182NQ87 A0A2A3EQW6 J9HZ80 O77168 A0A195BUL5 A0A0L7RB50 A0A0K8V3V4
Ontologies
GO
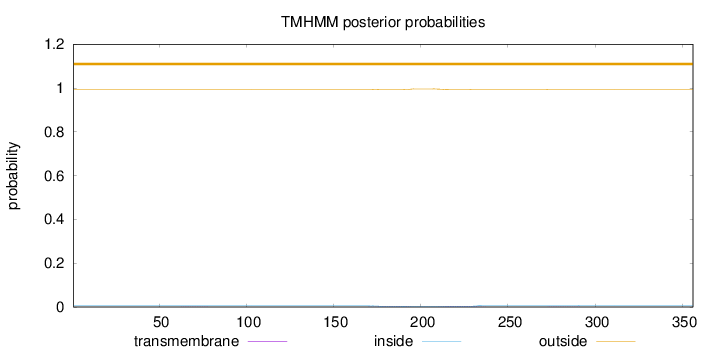
Topology
Subcellular location
Nucleus
Length:
356
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.15601
Exp number, first 60 AAs:
0.00021
Total prob of N-in:
0.00525
outside
1 - 356
Population Genetic Test Statistics
Pi
18.485688
Theta
14.747971
Tajima's D
0.215534
CLR
22.478623
CSRT
0.44412779361032
Interpretation
Uncertain
