Gene
KWMTBOMO06200
Pre Gene Modal
BGIBMGA001656
Annotation
PREDICTED:_zinc_finger_and_BTB_domain-containing_protein_41-like_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 2.584
Sequence
CDS
ATGTGCAACAAGGGCTACCCGACGAAGGAAGCGATGCAGGACCACTTCAACTACCAGCACCTGGGGAAGACGGCGCACAAGTGTCCCGTGTGCGAAAAGCCCATAGCCTCCAGGGCGAATGTTGTGAAGCACGTGATGAGGGTGCACGGCGAGAAGAAGGAGAAGCCGAGGAACCACGCCTGCGGGATGTGCAGGAAACGGTTCACTGACAAGAAGGCTCTGACTCAGCACGAGGTGATCCACTCCCGGGAGAGGCCGCTGACCTGCGACATCTGCCAACAAACCTTCAAGCAGAAGGCCTCTCTTTACACGCACAAGAAGAGGGTCCACAAAGTCGTGCCCAACAAGAAAGTCGTTGAATTCATTGAGGCGACGGCTTAG
Protein
MCNKGYPTKEAMQDHFNYQHLGKTAHKCPVCEKPIASRANVVKHVMRVHGEKKEKPRNHACGMCRKRFTDKKALTQHEVIHSRERPLTCDICQQTFKQKASLYTHKKRVHKVVPNKKVVEFIEATA
Summary
Uniprot
H9IWM4
S4NN42
A0A2A4J9S1
A0A212F8F4
A0A2H1VWX0
A0A1E1W6H5
+ More
A0A0N1IJT5 A0A0N1PHY6 A0A0N1IEI9 A0A2A4JCU5 A0A194Q532 A0A212F479 S4PHB1 A0A0L7KSP2 S4PRB7 A0A3S2NV19 A0A194Q4K3 A0A2W1BK83 A0A194QRA1 A0A2H1WUB2 S4PEZ1 A0A2J7Q4B9 A0A2L2YHF1 A0A0N0PBL1 A0A212FA07 A0A2A4J7P7 H9IWM9 A0A0N1IFW2 A0A087V0W8 A0A2A4JMU5 A0A0J7L1L6 A0A0N0PAK7 A0A3S2L580 A0A3B3ZBE0 A0A087TF09 A0A087UCD0 S4P3B4 A0A3S2NQN3 A0A087TG18 A0A0N1ICJ9 A0A0L8GHJ3 A0A087TUW7 A0A2P6KBF7 A0A3Q2G8Q1 A0A067R7K3 A0A2P8XD35 A0A2A4JBW6 A0A2H1WLQ9 A0A087V0W9 A0A0P5QFC0 A0A0P5R345 A0A0N8BUV5 A0A2W1B042 A0A2P8YGJ1 A0A131Y2W7 A0A087UA88 Q17MJ3 A0A0P5XLI7 A0A087UAK8 A0A0P5RY48 A0A0P5WGB2 A0A1B6EKM9 A0A0P5SJR4 A0A0P4YY80 A0A087TW41 A0A087T3Y6 A0A087U241 A0A0P5WK99 V4CBL5 A0A0P5E7I6 A0A0P5S9U1 A0A087V0T8 A0A0P5VJ40 A0A2P6KGY8 A0A067QTB7 A0A087V185 A0A2L2Z0J1 A0A0P5UK33 A0A3B3ZAU8 A0A087T206 A0A087V0T5 A0A3Q3BL37 A0A067R6D9 A0A087UDR9 R7VK40 A0A087U2E2 A0A087UAB3 V5HCN4 A0A232F202 A0A087V147 A0A0P6IXG9 A0A131Y1F7 A0A2L2YGM0 A0A147BPZ4 A0A2J7QR43 A0A3P9A1Y1
A0A0N1IJT5 A0A0N1PHY6 A0A0N1IEI9 A0A2A4JCU5 A0A194Q532 A0A212F479 S4PHB1 A0A0L7KSP2 S4PRB7 A0A3S2NV19 A0A194Q4K3 A0A2W1BK83 A0A194QRA1 A0A2H1WUB2 S4PEZ1 A0A2J7Q4B9 A0A2L2YHF1 A0A0N0PBL1 A0A212FA07 A0A2A4J7P7 H9IWM9 A0A0N1IFW2 A0A087V0W8 A0A2A4JMU5 A0A0J7L1L6 A0A0N0PAK7 A0A3S2L580 A0A3B3ZBE0 A0A087TF09 A0A087UCD0 S4P3B4 A0A3S2NQN3 A0A087TG18 A0A0N1ICJ9 A0A0L8GHJ3 A0A087TUW7 A0A2P6KBF7 A0A3Q2G8Q1 A0A067R7K3 A0A2P8XD35 A0A2A4JBW6 A0A2H1WLQ9 A0A087V0W9 A0A0P5QFC0 A0A0P5R345 A0A0N8BUV5 A0A2W1B042 A0A2P8YGJ1 A0A131Y2W7 A0A087UA88 Q17MJ3 A0A0P5XLI7 A0A087UAK8 A0A0P5RY48 A0A0P5WGB2 A0A1B6EKM9 A0A0P5SJR4 A0A0P4YY80 A0A087TW41 A0A087T3Y6 A0A087U241 A0A0P5WK99 V4CBL5 A0A0P5E7I6 A0A0P5S9U1 A0A087V0T8 A0A0P5VJ40 A0A2P6KGY8 A0A067QTB7 A0A087V185 A0A2L2Z0J1 A0A0P5UK33 A0A3B3ZAU8 A0A087T206 A0A087V0T5 A0A3Q3BL37 A0A067R6D9 A0A087UDR9 R7VK40 A0A087U2E2 A0A087UAB3 V5HCN4 A0A232F202 A0A087V147 A0A0P6IXG9 A0A131Y1F7 A0A2L2YGM0 A0A147BPZ4 A0A2J7QR43 A0A3P9A1Y1
Pubmed
EMBL
BABH01021167
GAIX01014096
JAA78464.1
NWSH01002274
PCG68711.1
AGBW02009729
+ More
OWR50026.1 ODYU01004954 SOQ45347.1 GDQN01008479 JAT82575.1 KQ459795 KPJ19918.1 KQ460555 KPJ14020.1 KQ458486 KPJ05869.1 NWSH01002068 PCG69260.1 KQ459463 KPJ00469.1 AGBW02010415 OWR48546.1 GAIX01000529 JAA92031.1 JTDY01006108 KOB66277.1 GAIX01000940 JAA91620.1 RSAL01000063 RVE49479.1 KPJ00468.1 KZ149995 PZC75449.1 KQ461198 KPJ06061.1 ODYU01010720 SOQ56004.1 GAIX01006840 JAA85720.1 NEVH01018385 PNF23428.1 IAAA01022758 IAAA01022759 LAA07467.1 KQ460969 KPJ10206.1 AGBW02009535 OWR50564.1 NWSH01002684 PCG67688.1 BABH01021155 KQ460140 KPJ17511.1 KL815794 KFM83257.1 NWSH01000953 PCG73395.1 LBMM01001240 KMQ96662.1 KQ458725 KPJ05348.1 RSAL01000142 RVE46032.1 KK114917 KFM63698.1 KK119191 KFM75019.1 GAIX01006514 JAA86046.1 RVE46029.1 KK115050 KFM64057.1 KPJ05382.1 KQ421769 KOF76466.1 KK116841 KFM68906.1 MWRG01017106 PRD23668.1 KK852889 KDR14291.1 PYGN01002835 PSN29910.1 PCG69256.1 ODYU01009516 SOQ54010.1 KFM83258.1 GDIQ01121621 JAL30105.1 GDIQ01233588 GDIQ01106132 GDIP01092879 JAL45594.1 JAM10836.1 GDIQ01142619 GDIP01124031 JAL09107.1 JAL79683.1 KZ150535 PZC70589.1 PYGN01000611 PSN43378.1 GEFM01002971 JAP72825.1 KK118960 KFM74277.1 CH477205 EAT47931.1 GDIP01070581 GDIQ01041460 JAM33134.1 JAN53277.1 KK119017 KFM74397.1 GDIQ01095521 JAL56205.1 GDIP01087188 JAM16527.1 GECZ01031294 JAS38475.1 GDIP01142587 JAL61127.1 GDIP01225320 JAI98081.1 KK117015 KFM69330.1 KK113288 KFM59825.1 KK117791 KFM71430.1 GDIP01097938 JAM05777.1 KB201038 ESO99264.1 GDIP01150523 JAJ72879.1 GDIP01142586 JAL61128.1 KL815034 KFM83227.1 GDIP01099278 JAM04437.1 MWRG01012206 PRD25592.1 KK852956 KDR13254.1 KL819331 KFM83374.1 IAAA01079269 LAA13240.1 GDIP01115822 JAL87892.1 KK113026 KFM59145.1 KL815010 KFM83224.1 KK852669 KDR18850.1 KK119373 KFM75508.1 AMQN01017480 KB293165 ELU16455.1 KK117836 KFM71531.1 KK118972 KFM74302.1 GANP01003193 JAB81275.1 NNAY01001291 OXU24467.1 KL818092 KFM83336.1 GDUN01000414 JAN95505.1 GEFM01003606 JAP72190.1 IAAA01027786 LAA07187.1 GEGO01002571 JAR92833.1 NEVH01011997 PNF31045.1
OWR50026.1 ODYU01004954 SOQ45347.1 GDQN01008479 JAT82575.1 KQ459795 KPJ19918.1 KQ460555 KPJ14020.1 KQ458486 KPJ05869.1 NWSH01002068 PCG69260.1 KQ459463 KPJ00469.1 AGBW02010415 OWR48546.1 GAIX01000529 JAA92031.1 JTDY01006108 KOB66277.1 GAIX01000940 JAA91620.1 RSAL01000063 RVE49479.1 KPJ00468.1 KZ149995 PZC75449.1 KQ461198 KPJ06061.1 ODYU01010720 SOQ56004.1 GAIX01006840 JAA85720.1 NEVH01018385 PNF23428.1 IAAA01022758 IAAA01022759 LAA07467.1 KQ460969 KPJ10206.1 AGBW02009535 OWR50564.1 NWSH01002684 PCG67688.1 BABH01021155 KQ460140 KPJ17511.1 KL815794 KFM83257.1 NWSH01000953 PCG73395.1 LBMM01001240 KMQ96662.1 KQ458725 KPJ05348.1 RSAL01000142 RVE46032.1 KK114917 KFM63698.1 KK119191 KFM75019.1 GAIX01006514 JAA86046.1 RVE46029.1 KK115050 KFM64057.1 KPJ05382.1 KQ421769 KOF76466.1 KK116841 KFM68906.1 MWRG01017106 PRD23668.1 KK852889 KDR14291.1 PYGN01002835 PSN29910.1 PCG69256.1 ODYU01009516 SOQ54010.1 KFM83258.1 GDIQ01121621 JAL30105.1 GDIQ01233588 GDIQ01106132 GDIP01092879 JAL45594.1 JAM10836.1 GDIQ01142619 GDIP01124031 JAL09107.1 JAL79683.1 KZ150535 PZC70589.1 PYGN01000611 PSN43378.1 GEFM01002971 JAP72825.1 KK118960 KFM74277.1 CH477205 EAT47931.1 GDIP01070581 GDIQ01041460 JAM33134.1 JAN53277.1 KK119017 KFM74397.1 GDIQ01095521 JAL56205.1 GDIP01087188 JAM16527.1 GECZ01031294 JAS38475.1 GDIP01142587 JAL61127.1 GDIP01225320 JAI98081.1 KK117015 KFM69330.1 KK113288 KFM59825.1 KK117791 KFM71430.1 GDIP01097938 JAM05777.1 KB201038 ESO99264.1 GDIP01150523 JAJ72879.1 GDIP01142586 JAL61128.1 KL815034 KFM83227.1 GDIP01099278 JAM04437.1 MWRG01012206 PRD25592.1 KK852956 KDR13254.1 KL819331 KFM83374.1 IAAA01079269 LAA13240.1 GDIP01115822 JAL87892.1 KK113026 KFM59145.1 KL815010 KFM83224.1 KK852669 KDR18850.1 KK119373 KFM75508.1 AMQN01017480 KB293165 ELU16455.1 KK117836 KFM71531.1 KK118972 KFM74302.1 GANP01003193 JAB81275.1 NNAY01001291 OXU24467.1 KL818092 KFM83336.1 GDUN01000414 JAN95505.1 GEFM01003606 JAP72190.1 IAAA01027786 LAA07187.1 GEGO01002571 JAR92833.1 NEVH01011997 PNF31045.1
Proteomes
Interpro
SUPFAM
SSF57667
SSF57667
ProteinModelPortal
H9IWM4
S4NN42
A0A2A4J9S1
A0A212F8F4
A0A2H1VWX0
A0A1E1W6H5
+ More
A0A0N1IJT5 A0A0N1PHY6 A0A0N1IEI9 A0A2A4JCU5 A0A194Q532 A0A212F479 S4PHB1 A0A0L7KSP2 S4PRB7 A0A3S2NV19 A0A194Q4K3 A0A2W1BK83 A0A194QRA1 A0A2H1WUB2 S4PEZ1 A0A2J7Q4B9 A0A2L2YHF1 A0A0N0PBL1 A0A212FA07 A0A2A4J7P7 H9IWM9 A0A0N1IFW2 A0A087V0W8 A0A2A4JMU5 A0A0J7L1L6 A0A0N0PAK7 A0A3S2L580 A0A3B3ZBE0 A0A087TF09 A0A087UCD0 S4P3B4 A0A3S2NQN3 A0A087TG18 A0A0N1ICJ9 A0A0L8GHJ3 A0A087TUW7 A0A2P6KBF7 A0A3Q2G8Q1 A0A067R7K3 A0A2P8XD35 A0A2A4JBW6 A0A2H1WLQ9 A0A087V0W9 A0A0P5QFC0 A0A0P5R345 A0A0N8BUV5 A0A2W1B042 A0A2P8YGJ1 A0A131Y2W7 A0A087UA88 Q17MJ3 A0A0P5XLI7 A0A087UAK8 A0A0P5RY48 A0A0P5WGB2 A0A1B6EKM9 A0A0P5SJR4 A0A0P4YY80 A0A087TW41 A0A087T3Y6 A0A087U241 A0A0P5WK99 V4CBL5 A0A0P5E7I6 A0A0P5S9U1 A0A087V0T8 A0A0P5VJ40 A0A2P6KGY8 A0A067QTB7 A0A087V185 A0A2L2Z0J1 A0A0P5UK33 A0A3B3ZAU8 A0A087T206 A0A087V0T5 A0A3Q3BL37 A0A067R6D9 A0A087UDR9 R7VK40 A0A087U2E2 A0A087UAB3 V5HCN4 A0A232F202 A0A087V147 A0A0P6IXG9 A0A131Y1F7 A0A2L2YGM0 A0A147BPZ4 A0A2J7QR43 A0A3P9A1Y1
A0A0N1IJT5 A0A0N1PHY6 A0A0N1IEI9 A0A2A4JCU5 A0A194Q532 A0A212F479 S4PHB1 A0A0L7KSP2 S4PRB7 A0A3S2NV19 A0A194Q4K3 A0A2W1BK83 A0A194QRA1 A0A2H1WUB2 S4PEZ1 A0A2J7Q4B9 A0A2L2YHF1 A0A0N0PBL1 A0A212FA07 A0A2A4J7P7 H9IWM9 A0A0N1IFW2 A0A087V0W8 A0A2A4JMU5 A0A0J7L1L6 A0A0N0PAK7 A0A3S2L580 A0A3B3ZBE0 A0A087TF09 A0A087UCD0 S4P3B4 A0A3S2NQN3 A0A087TG18 A0A0N1ICJ9 A0A0L8GHJ3 A0A087TUW7 A0A2P6KBF7 A0A3Q2G8Q1 A0A067R7K3 A0A2P8XD35 A0A2A4JBW6 A0A2H1WLQ9 A0A087V0W9 A0A0P5QFC0 A0A0P5R345 A0A0N8BUV5 A0A2W1B042 A0A2P8YGJ1 A0A131Y2W7 A0A087UA88 Q17MJ3 A0A0P5XLI7 A0A087UAK8 A0A0P5RY48 A0A0P5WGB2 A0A1B6EKM9 A0A0P5SJR4 A0A0P4YY80 A0A087TW41 A0A087T3Y6 A0A087U241 A0A0P5WK99 V4CBL5 A0A0P5E7I6 A0A0P5S9U1 A0A087V0T8 A0A0P5VJ40 A0A2P6KGY8 A0A067QTB7 A0A087V185 A0A2L2Z0J1 A0A0P5UK33 A0A3B3ZAU8 A0A087T206 A0A087V0T5 A0A3Q3BL37 A0A067R6D9 A0A087UDR9 R7VK40 A0A087U2E2 A0A087UAB3 V5HCN4 A0A232F202 A0A087V147 A0A0P6IXG9 A0A131Y1F7 A0A2L2YGM0 A0A147BPZ4 A0A2J7QR43 A0A3P9A1Y1
PDB
2I13
E-value=5.75938e-07,
Score=121
Ontologies
GO
Topology
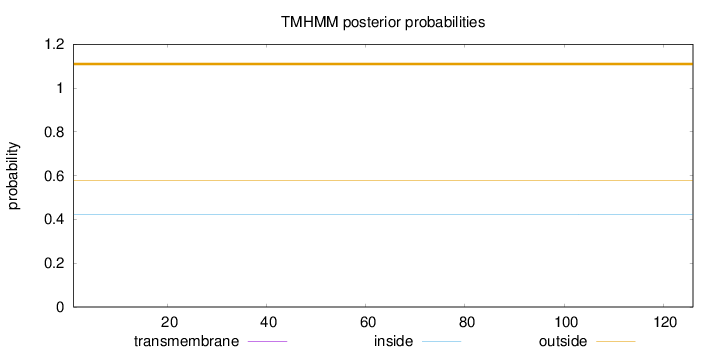
Length:
126
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.42136
outside
1 - 126
Population Genetic Test Statistics
Pi
156.819073
Theta
139.870282
Tajima's D
1.183047
CLR
0
CSRT
0.710814459277036
Interpretation
Uncertain
