Gene
KWMTBOMO06192
Pre Gene Modal
BGIBMGA001660
Annotation
PREDICTED:_zinc_finger_protein_62-like_isoform_X3_[Bombyx_mori]
Full name
Zinc finger protein 57
+ More
Zinc finger protein
Zinc finger protein
Alternative Name
Zinc finger protein 424
Transcription factor
Location in the cell
Nuclear Reliability : 4.522
Sequence
CDS
ATGAAAGAGGATGGCTGGAACCTTGAGCTTGTCGGCGGACGTGAAGAGATGAAAAAGGATGGCTGGATCCTTGAGCTTGTCGGCGGGATCAGATTGATGGTGATAGAGTTATCTGACGCCGAGCTAGGGGCTGAGAGGGAGAGGGCCATCCGCGACCCTAGATACACAAAGAAGTCGTACAAATGTCAGAAATGTATAATCTCATACGACCACGCCATTAACTTGGACAGGCATTTGACACTCAGCCATAATGAGAATAACGCACACGTGTGCGACGTATGCGAATGTACTTACGACACAGAATCGAAAGTCAGAACTCACACGAAGAGACATTATTTGAGGTACAAGTGTATGGAATGCGATAAGTTGTACCAGGACAAACAAACAGCCGTTGTGCACTACAAAGAAGCACATACGGTCAATCGGAAGATATACAGATGCGATCTGTCGCACTGTCGCTTCGAGACTGCCTCTCACAGAAGCTACAAATATCATCAAGCGAAACACAATCAAGTTAAATGCTCTTTATGCGACGGAATGTACTTGCAGGGGACCCAATTGAGGATGCATATCAACACCGTCCACAAAAACGCATCGCAATTCAAGTGCGAGTCGTGCGATAAGGTCTACAAGCACCGGGCAGGGTTGCGCGCGCACGTGAGGTCGGCACACGAAGCGAACAACGAGACGTCTCATTACTGCGGCCCTTGTTGCAAATTCTTCAGCACCGCCTCCACGCTGGCCCAACATCTGATCGAACATTCGAAGCATGTCGGCAAAAAATACGTTTGCGATGATTGCTCAAAGAGATACTCCACGAAGCGCTTCCTGAGACAACACATAGAAACGGCACATTTGAAGCTCAAACCCGATTACAAGTGCACGATTTGCGATAAGATCTTCAATACGTCGTTCACGCTCAGAAGACACGTGGAGTTTAAACACGAGAAAAAAAAGTTGCCTCGCGATAAAATCTGCGAACATTGCGGACGCGGGTTCATGACGAACAAAATACTGAGTTCGCATATACGCACGCACACCGGCGAACGGCCGTACGTGTGCCAGCTCTGCGGCGCGACTTTTGGCCACTCGGGCGCGATGTACACCCATAAAAGGTTACTGCACGGCTTAAAAAAACGTAAAGGAAGCTAG
Protein
MKEDGWNLELVGGREEMKKDGWILELVGGIRLMVIELSDAELGAERERAIRDPRYTKKSYKCQKCIISYDHAINLDRHLTLSHNENNAHVCDVCECTYDTESKVRTHTKRHYLRYKCMECDKLYQDKQTAVVHYKEAHTVNRKIYRCDLSHCRFETASHRSYKYHQAKHNQVKCSLCDGMYLQGTQLRMHINTVHKNASQFKCESCDKVYKHRAGLRAHVRSAHEANNETSHYCGPCCKFFSTASTLAQHLIEHSKHVGKKYVCDDCSKRYSTKRFLRQHIETAHLKLKPDYKCTICDKIFNTSFTLRRHVEFKHEKKKLPRDKICEHCGRGFMTNKILSSHIRTHTGERPYVCQLCGATFGHSGAMYTHKRLLHGLKKRKGS
Summary
Similarity
Belongs to the krueppel C2H2-type zinc-finger protein family.
Keywords
Complete proteome
DNA-binding
Metal-binding
Nucleus
Polymorphism
Reference proteome
Repeat
Transcription
Transcription regulation
Zinc
Zinc-finger
Feature
chain Zinc finger protein 57
sequence variant In dbSNP:rs2288958.
sequence variant In dbSNP:rs2288958.
Uniprot
A0A2W1BK83
A0A3S2L580
A0A194QRA1
A0A212FA07
A0A0N1IE69
A0A0N0PE54
+ More
A0A0N1IGS5 A0A2A4JMU5 A0A3S2NQN3 H9IWZ4 A0A2A4JGB7 A0A0N0PBL1 A0A212F827 A0A3S2NF42 A0A2H1W7Z1 A0A3S2NV19 A0A2H1WUB2 X1WX70 A0A0N1ICJ9 S4PJM9 A0A2A4K9P1 A0A2I3GCI3 A0A2J8J2W2 A0A2I3HD63 H2REK2 H2QEX6 A0A087WUM7 A0A2J8J2W0 A0A2K5VLF6 B4DXX0 A5HJR3 Q68EA5 A0A2K5VLD9 G3V131 A0A2K5VL69 A0A2K6BUY8 A0A2K6BUZ2 A0A1D5RH92 A0A2K6BV28 A0A2K6BUZ9 A0A2K6BV06 A0A2Y9THY0 J9JWR5 A0A2I3LRP3 A0A212F8F4 A0A2K5NKU0 A0A2K5NKT0 A0A0A0MWA1 A0A2K5NKF4 J9KG92 A0A2A4K4S3 G3RMR5 A0A2P6KCU3 A0A2K6P4J1 A0A2K6N5E7 G3QTW4 A0A2P6KDB3 A0A0D9RGB2 A0A2K6P4I3 A0A2K5XIM1 A0A2K5XIM7 A0A2K6N5E5 V4CF43 A0A1S3PMQ0 A0A1S3QLL2 A0A3S2L825 A0A2P6K6Q8 A0A2K5HFH4 C3ZT30 C4A0V9 A0A1E1WCP9 A0A2K5HFE9 A0A2P6JVG6 J9JNC1 A0A2P6K6P9 W4X9H3 A0A2P6K0D1 A0A139WK46 F1MJ44 G7NLD6 A0A2K6LD93 A0A0L8FSW4 A0A2P6K7A6 A0A2P6K1M6 A0A3P8SWG4 G3SB27 G3QWE7 A0A2P6K7N8 A0A2Y9RY84 A0A2I3M0J2 G7PZG4 H2NY78 H2QFW5 W4YY05 A0A2J8LSD7
A0A0N1IGS5 A0A2A4JMU5 A0A3S2NQN3 H9IWZ4 A0A2A4JGB7 A0A0N0PBL1 A0A212F827 A0A3S2NF42 A0A2H1W7Z1 A0A3S2NV19 A0A2H1WUB2 X1WX70 A0A0N1ICJ9 S4PJM9 A0A2A4K9P1 A0A2I3GCI3 A0A2J8J2W2 A0A2I3HD63 H2REK2 H2QEX6 A0A087WUM7 A0A2J8J2W0 A0A2K5VLF6 B4DXX0 A5HJR3 Q68EA5 A0A2K5VLD9 G3V131 A0A2K5VL69 A0A2K6BUY8 A0A2K6BUZ2 A0A1D5RH92 A0A2K6BV28 A0A2K6BUZ9 A0A2K6BV06 A0A2Y9THY0 J9JWR5 A0A2I3LRP3 A0A212F8F4 A0A2K5NKU0 A0A2K5NKT0 A0A0A0MWA1 A0A2K5NKF4 J9KG92 A0A2A4K4S3 G3RMR5 A0A2P6KCU3 A0A2K6P4J1 A0A2K6N5E7 G3QTW4 A0A2P6KDB3 A0A0D9RGB2 A0A2K6P4I3 A0A2K5XIM1 A0A2K5XIM7 A0A2K6N5E5 V4CF43 A0A1S3PMQ0 A0A1S3QLL2 A0A3S2L825 A0A2P6K6Q8 A0A2K5HFH4 C3ZT30 C4A0V9 A0A1E1WCP9 A0A2K5HFE9 A0A2P6JVG6 J9JNC1 A0A2P6K6P9 W4X9H3 A0A2P6K0D1 A0A139WK46 F1MJ44 G7NLD6 A0A2K6LD93 A0A0L8FSW4 A0A2P6K7A6 A0A2P6K1M6 A0A3P8SWG4 G3SB27 G3QWE7 A0A2P6K7N8 A0A2Y9RY84 A0A2I3M0J2 G7PZG4 H2NY78 H2QFW5 W4YY05 A0A2J8LSD7
EC Number
2.1.1.-
Pubmed
EMBL
KZ149995
PZC75449.1
RSAL01000142
RVE46032.1
KQ461198
KPJ06061.1
+ More
AGBW02009535 OWR50564.1 KQ458725 KPJ05352.1 KQ459983 KPJ19122.1 KQ460400 KPJ14980.1 NWSH01000953 PCG73395.1 RVE46029.1 BABH01021154 NWSH01001510 PCG71021.1 KQ460969 KPJ10206.1 AGBW02009801 OWR49895.1 RSAL01000063 RVE49477.1 ODYU01006917 SOQ49191.1 RVE49479.1 ODYU01010720 SOQ56004.1 ABLF02038592 KPJ05382.1 GAIX01004880 JAA87680.1 NWSH01000010 PCG80927.1 ADFV01098915 ADFV01098916 ADFV01098917 ADFV01098918 ADFV01098919 NBAG03000531 PNI17105.1 AACZ04058062 PNI17104.1 AC119403 PNI17106.1 AQIA01033538 AQIA01033539 AQIA01033540 AQIA01033541 AK302165 BAG63532.1 EF534355 AK314848 CH471139 ABQ10558.1 BAG37365.1 EAW69355.1 BX537601 BC028974 EAW69356.1 JSUE03020218 JSUE03020219 JSUE03020220 JSUE03020221 ABLF02038588 ABLF02038606 ABLF02038607 AHZZ02013168 AHZZ02013169 AHZZ02013170 AHZZ02013171 AGBW02009729 OWR50026.1 ABLF02037187 NWSH01000147 PCG79069.1 CABD030110760 MWRG01015819 PRD24148.1 MWRG01015375 PRD24313.1 AQIB01150271 KB200701 ESP00620.1 RSAL01000885 RVE41075.1 MWRG01023322 PRD22014.1 GG666676 EEN44236.1 GG666831 EEN41572.1 GDQN01006290 GDQN01003686 JAT84764.1 JAT87368.1 MWRG01130324 PRD18060.1 ABLF02025528 MWRG01023406 PRD21999.1 AAGJ04106031 AAGJ04106032 AAGJ04106033 MWRG01051860 PRD19789.1 KQ971332 KYB28274.1 CM001271 EHH29861.1 KQ426791 KOF67769.1 MWRG01022237 PRD22226.1 MWRG01042629 PRD20213.1 CABD030111734 CABD030112448 CABD030112449 CABD030112450 CABD030112451 CABD030112452 CABD030112453 CABD030112454 MWRG01021789 PRD22340.1 AHZZ02013870 AHZZ02013871 AHZZ02013872 AHZZ02013873 AHZZ02013874 AHZZ02013875 AHZZ02013876 AHZZ02013877 AHZZ02013878 CM001294 EHH59234.1 ABGA01347901 ABGA01347902 AACZ04030926 AAGJ04134332 AAGJ04134333 AAGJ04134334 AAGJ04134335 NBAG03000278 PNI50190.1
AGBW02009535 OWR50564.1 KQ458725 KPJ05352.1 KQ459983 KPJ19122.1 KQ460400 KPJ14980.1 NWSH01000953 PCG73395.1 RVE46029.1 BABH01021154 NWSH01001510 PCG71021.1 KQ460969 KPJ10206.1 AGBW02009801 OWR49895.1 RSAL01000063 RVE49477.1 ODYU01006917 SOQ49191.1 RVE49479.1 ODYU01010720 SOQ56004.1 ABLF02038592 KPJ05382.1 GAIX01004880 JAA87680.1 NWSH01000010 PCG80927.1 ADFV01098915 ADFV01098916 ADFV01098917 ADFV01098918 ADFV01098919 NBAG03000531 PNI17105.1 AACZ04058062 PNI17104.1 AC119403 PNI17106.1 AQIA01033538 AQIA01033539 AQIA01033540 AQIA01033541 AK302165 BAG63532.1 EF534355 AK314848 CH471139 ABQ10558.1 BAG37365.1 EAW69355.1 BX537601 BC028974 EAW69356.1 JSUE03020218 JSUE03020219 JSUE03020220 JSUE03020221 ABLF02038588 ABLF02038606 ABLF02038607 AHZZ02013168 AHZZ02013169 AHZZ02013170 AHZZ02013171 AGBW02009729 OWR50026.1 ABLF02037187 NWSH01000147 PCG79069.1 CABD030110760 MWRG01015819 PRD24148.1 MWRG01015375 PRD24313.1 AQIB01150271 KB200701 ESP00620.1 RSAL01000885 RVE41075.1 MWRG01023322 PRD22014.1 GG666676 EEN44236.1 GG666831 EEN41572.1 GDQN01006290 GDQN01003686 JAT84764.1 JAT87368.1 MWRG01130324 PRD18060.1 ABLF02025528 MWRG01023406 PRD21999.1 AAGJ04106031 AAGJ04106032 AAGJ04106033 MWRG01051860 PRD19789.1 KQ971332 KYB28274.1 CM001271 EHH29861.1 KQ426791 KOF67769.1 MWRG01022237 PRD22226.1 MWRG01042629 PRD20213.1 CABD030111734 CABD030112448 CABD030112449 CABD030112450 CABD030112451 CABD030112452 CABD030112453 CABD030112454 MWRG01021789 PRD22340.1 AHZZ02013870 AHZZ02013871 AHZZ02013872 AHZZ02013873 AHZZ02013874 AHZZ02013875 AHZZ02013876 AHZZ02013877 AHZZ02013878 CM001294 EHH59234.1 ABGA01347901 ABGA01347902 AACZ04030926 AAGJ04134332 AAGJ04134333 AAGJ04134334 AAGJ04134335 NBAG03000278 PNI50190.1
Proteomes
UP000283053
UP000053240
UP000007151
UP000053268
UP000218220
UP000005204
+ More
UP000007819 UP000001073 UP000002277 UP000005640 UP000233100 UP000233120 UP000006718 UP000028761 UP000233060 UP000001519 UP000233200 UP000233180 UP000029965 UP000233140 UP000030746 UP000087266 UP000233080 UP000001554 UP000007110 UP000007266 UP000009136 UP000053454 UP000265080 UP000248480 UP000009130 UP000001595
UP000007819 UP000001073 UP000002277 UP000005640 UP000233100 UP000233120 UP000006718 UP000028761 UP000233060 UP000001519 UP000233200 UP000233180 UP000029965 UP000233140 UP000030746 UP000087266 UP000233080 UP000001554 UP000007110 UP000007266 UP000009136 UP000053454 UP000265080 UP000248480 UP000009130 UP000001595
Pfam
Interpro
IPR012934
Znf_AD
+ More
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR022755 Znf_C2H2_jaz
IPR003604 Matrin/U1-like-C_Znf_C2H2
IPR017114 YY-1-like
IPR036051 KRAB_dom_sf
IPR001909 KRAB
IPR008598 Di19_Zn_binding_dom
IPR003656 Znf_BED
IPR041661 ZN622/Rei1/Reh1_Znf-C2H2
IPR001965 Znf_PHD
IPR025398 DUF4371
IPR017125 Znf_PRDM5-like
IPR011017 TRASH_dom
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR022755 Znf_C2H2_jaz
IPR003604 Matrin/U1-like-C_Znf_C2H2
IPR017114 YY-1-like
IPR036051 KRAB_dom_sf
IPR001909 KRAB
IPR008598 Di19_Zn_binding_dom
IPR003656 Znf_BED
IPR041661 ZN622/Rei1/Reh1_Znf-C2H2
IPR001965 Znf_PHD
IPR025398 DUF4371
IPR017125 Znf_PRDM5-like
IPR011017 TRASH_dom
ProteinModelPortal
A0A2W1BK83
A0A3S2L580
A0A194QRA1
A0A212FA07
A0A0N1IE69
A0A0N0PE54
+ More
A0A0N1IGS5 A0A2A4JMU5 A0A3S2NQN3 H9IWZ4 A0A2A4JGB7 A0A0N0PBL1 A0A212F827 A0A3S2NF42 A0A2H1W7Z1 A0A3S2NV19 A0A2H1WUB2 X1WX70 A0A0N1ICJ9 S4PJM9 A0A2A4K9P1 A0A2I3GCI3 A0A2J8J2W2 A0A2I3HD63 H2REK2 H2QEX6 A0A087WUM7 A0A2J8J2W0 A0A2K5VLF6 B4DXX0 A5HJR3 Q68EA5 A0A2K5VLD9 G3V131 A0A2K5VL69 A0A2K6BUY8 A0A2K6BUZ2 A0A1D5RH92 A0A2K6BV28 A0A2K6BUZ9 A0A2K6BV06 A0A2Y9THY0 J9JWR5 A0A2I3LRP3 A0A212F8F4 A0A2K5NKU0 A0A2K5NKT0 A0A0A0MWA1 A0A2K5NKF4 J9KG92 A0A2A4K4S3 G3RMR5 A0A2P6KCU3 A0A2K6P4J1 A0A2K6N5E7 G3QTW4 A0A2P6KDB3 A0A0D9RGB2 A0A2K6P4I3 A0A2K5XIM1 A0A2K5XIM7 A0A2K6N5E5 V4CF43 A0A1S3PMQ0 A0A1S3QLL2 A0A3S2L825 A0A2P6K6Q8 A0A2K5HFH4 C3ZT30 C4A0V9 A0A1E1WCP9 A0A2K5HFE9 A0A2P6JVG6 J9JNC1 A0A2P6K6P9 W4X9H3 A0A2P6K0D1 A0A139WK46 F1MJ44 G7NLD6 A0A2K6LD93 A0A0L8FSW4 A0A2P6K7A6 A0A2P6K1M6 A0A3P8SWG4 G3SB27 G3QWE7 A0A2P6K7N8 A0A2Y9RY84 A0A2I3M0J2 G7PZG4 H2NY78 H2QFW5 W4YY05 A0A2J8LSD7
A0A0N1IGS5 A0A2A4JMU5 A0A3S2NQN3 H9IWZ4 A0A2A4JGB7 A0A0N0PBL1 A0A212F827 A0A3S2NF42 A0A2H1W7Z1 A0A3S2NV19 A0A2H1WUB2 X1WX70 A0A0N1ICJ9 S4PJM9 A0A2A4K9P1 A0A2I3GCI3 A0A2J8J2W2 A0A2I3HD63 H2REK2 H2QEX6 A0A087WUM7 A0A2J8J2W0 A0A2K5VLF6 B4DXX0 A5HJR3 Q68EA5 A0A2K5VLD9 G3V131 A0A2K5VL69 A0A2K6BUY8 A0A2K6BUZ2 A0A1D5RH92 A0A2K6BV28 A0A2K6BUZ9 A0A2K6BV06 A0A2Y9THY0 J9JWR5 A0A2I3LRP3 A0A212F8F4 A0A2K5NKU0 A0A2K5NKT0 A0A0A0MWA1 A0A2K5NKF4 J9KG92 A0A2A4K4S3 G3RMR5 A0A2P6KCU3 A0A2K6P4J1 A0A2K6N5E7 G3QTW4 A0A2P6KDB3 A0A0D9RGB2 A0A2K6P4I3 A0A2K5XIM1 A0A2K5XIM7 A0A2K6N5E5 V4CF43 A0A1S3PMQ0 A0A1S3QLL2 A0A3S2L825 A0A2P6K6Q8 A0A2K5HFH4 C3ZT30 C4A0V9 A0A1E1WCP9 A0A2K5HFE9 A0A2P6JVG6 J9JNC1 A0A2P6K6P9 W4X9H3 A0A2P6K0D1 A0A139WK46 F1MJ44 G7NLD6 A0A2K6LD93 A0A0L8FSW4 A0A2P6K7A6 A0A2P6K1M6 A0A3P8SWG4 G3SB27 G3QWE7 A0A2P6K7N8 A0A2Y9RY84 A0A2I3M0J2 G7PZG4 H2NY78 H2QFW5 W4YY05 A0A2J8LSD7
PDB
5V3M
E-value=8.28128e-20,
Score=239
Ontologies
GO
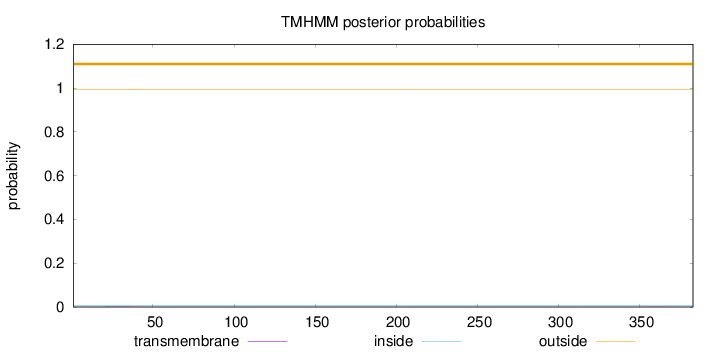
Topology
Subcellular location
Nucleus
Length:
383
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03973
Exp number, first 60 AAs:
0.03024
Total prob of N-in:
0.00814
outside
1 - 383
Population Genetic Test Statistics
Pi
275.851547
Theta
169.389773
Tajima's D
1.809195
CLR
0.012638
CSRT
0.846257687115644
Interpretation
Uncertain
