Gene
KWMTBOMO06189
Pre Gene Modal
BGIBMGA001777
Annotation
PREDICTED:_zinc_finger_protein_708-like_isoform_X1_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 4.236
Sequence
CDS
ATGAAGACAGTTAAATTAGAAATTTCAGTTGACACAGTGTGCCATGGGTGTTTAAGTACCGATAGGCAAACAAGCACTTCGAGAGATACATCTGAATTATATTTACGTTTATTAGAAGACGGTTTATCCTCTGAAAATAAGGATGTGGCTCTCTGTTGGGAATGTACAGCAATCTTCAGGAAGATCCAGAGGTTTCAGAAGCAAATTAAAAACGCTCAAATCTCTCTTCTCTTATACCAATTGAATACAAGGACACCGTTATCAAAATTAGATACCGTAATCAAAAGTGTATACGATCTGGAGTGTGTACACGAAGAGAAAGCGGCACAAATAACCGAAGAAGGTAACTCGATAAAAATAGAAGCGGAAACGGAAGTCGCACTGCCCGCAAAAGAAGAATTAGACGAATATGACACTATAACGTACGAGGAGGACTACGCTGATGATGCGTGTATGAAAAGCATAATTGAAAACGAACAGAGAGCCAGCAAAATGCCAAGTAAGAGCGATATCAAACCCATGTCCATAAAGCAATACGAGAAATATGCAGTTTTCTCGAAGCAGAACGAGAAAGTTAATTCTGGTGACCTGAAAAAATACTATAAGAACGTTTTCTTGAGTGACGGTGAGATAGAGGAATATTTGAACACTAGATTAAGGAGGGATAAGAGCTTCCATCGTTTGAATTACAAGTGCGAGGATTGTGTACTCGGATACAAGGATAAGAGGGACTGGAATCGGCATAATGCTCTTCATCATAACACTTCGACCGGCCCCTACAAGTGCAGCGAGTGCAAAACGAAATGCCAAACCATCGATGTACTCGCCCAGCATTGGATAACTCACACGAAGGCGCTGCAGTGCGTCATCTGCGGCGACCTGCACAGGTCCCTGGGTGAGATAAGGAAGCACGTGAACAGGGCGCACACGGGCGTGTTCACGTGCAAGGAGTGCGGGGACCACAGCAGAACACTAAGGGAGTTCAGTCAGCATTACAAAAGCAAACACGAGAAGCTGGTATGTGATCATTGCGGCAAAGGTTTTTACAAGAAGCGCGTCCTAGAAAGTCATATGAGGAGGAACCATTTACCAGCCAAATGCGAGGTCTGCGGCCGTCAGTACTCTCTGTACCACACCCTGGAGGTTCACCTGCGGACTGTCCATCCTCACCTCATGAACGGGGCCTACAATAGGGACGCCTCGTACTGCGTCGAGTGCGATAGACAGTACCCAAGCGTCTACAAATACAGGAAGCATTTGAAACAGTCGGTCAGGCATACCCCGAAAAAGAAAGTAAGGATACCGTGCCCCGAATGCGGAAAAGTCTTCACGCGTACGAACTACATGAACAATCACTACAGGCTGTTCCATTCCAAGGACACGAAGCACTATTGTCAGCTTTGTAACAAGCTCTTCGTCACCGGCTACGCGGCCAGGAAACACAAAGAGTTCGTTCACGACAAACAAACTTTGCCCAAAAACAAGATCTGCGATATATGCGGCCGCGGCTTTAGCACGAACAGAATTCTAACCAATCACAGGAGGACACACACCGGAGAGAGACCGTACAAATGCCCGCACTGTACCGCCGCGTTCGCCCAAAGCACCGCCATGCACACGCACATGAAATCGCAACACAAACACGTAATGCCTCTACAGATCCCCGTCCACCAGATGGCGTGA
Protein
MKTVKLEISVDTVCHGCLSTDRQTSTSRDTSELYLRLLEDGLSSENKDVALCWECTAIFRKIQRFQKQIKNAQISLLLYQLNTRTPLSKLDTVIKSVYDLECVHEEKAAQITEEGNSIKIEAETEVALPAKEELDEYDTITYEEDYADDACMKSIIENEQRASKMPSKSDIKPMSIKQYEKYAVFSKQNEKVNSGDLKKYYKNVFLSDGEIEEYLNTRLRRDKSFHRLNYKCEDCVLGYKDKRDWNRHNALHHNTSTGPYKCSECKTKCQTIDVLAQHWITHTKALQCVICGDLHRSLGEIRKHVNRAHTGVFTCKECGDHSRTLREFSQHYKSKHEKLVCDHCGKGFYKKRVLESHMRRNHLPAKCEVCGRQYSLYHTLEVHLRTVHPHLMNGAYNRDASYCVECDRQYPSVYKYRKHLKQSVRHTPKKKVRIPCPECGKVFTRTNYMNNHYRLFHSKDTKHYCQLCNKLFVTGYAARKHKEFVHDKQTLPKNKICDICGRGFSTNRILTNHRRTHTGERPYKCPHCTAAFAQSTAMHTHMKSQHKHVMPLQIPVHQMA
Summary
Uniprot
H9IWZ4
A0A2A4K9P1
A0A3S2NW05
A0A2A4KAB9
A0A2A4JMU5
A0A3S2THA9
+ More
A0A3S2NF42 A0A2W1BK83 A0A0N0PBL1 A0A3S2NV19 A0A2H1WUB2 A0A0N1ICJ9 A0A212FHB4 A0A2A4JGB7 A0A1E1WCP9 A0A212F827 A0A2H1W7Z1 A0A087YR65 A0A3B3UVZ4 A0A3B3YZ32 A0A0N1IEI9 A0A0N0PE54 A0A0N1IFW2 C3ZPQ0 A0A3B3UVG3 A0A212F479 A0A3Q3IRR6 L7MZU6 W5P8U9
A0A3S2NF42 A0A2W1BK83 A0A0N0PBL1 A0A3S2NV19 A0A2H1WUB2 A0A0N1ICJ9 A0A212FHB4 A0A2A4JGB7 A0A1E1WCP9 A0A212F827 A0A2H1W7Z1 A0A087YR65 A0A3B3UVZ4 A0A3B3YZ32 A0A0N1IEI9 A0A0N0PE54 A0A0N1IFW2 C3ZPQ0 A0A3B3UVG3 A0A212F479 A0A3Q3IRR6 L7MZU6 W5P8U9
EMBL
BABH01021154
NWSH01000010
PCG80927.1
RSAL01000142
RVE46036.1
PCG80926.1
+ More
NWSH01000953 PCG73395.1 RVE46037.1 RSAL01000063 RVE49477.1 KZ149995 PZC75449.1 KQ460969 KPJ10206.1 RVE49479.1 ODYU01010720 SOQ56004.1 KQ458725 KPJ05382.1 AGBW02008515 OWR53125.1 NWSH01001510 PCG71021.1 GDQN01006290 GDQN01003686 JAT84764.1 JAT87368.1 AGBW02009801 OWR49895.1 ODYU01006917 SOQ49191.1 AYCK01013004 KQ458486 KPJ05869.1 KQ459983 KPJ19122.1 KQ460140 KPJ17511.1 GG666659 EEN45471.1 AGBW02010415 OWR48546.1 AAWZ02035144 AAWZ02035145 AMGL01047138
NWSH01000953 PCG73395.1 RVE46037.1 RSAL01000063 RVE49477.1 KZ149995 PZC75449.1 KQ460969 KPJ10206.1 RVE49479.1 ODYU01010720 SOQ56004.1 KQ458725 KPJ05382.1 AGBW02008515 OWR53125.1 NWSH01001510 PCG71021.1 GDQN01006290 GDQN01003686 JAT84764.1 JAT87368.1 AGBW02009801 OWR49895.1 ODYU01006917 SOQ49191.1 AYCK01013004 KQ458486 KPJ05869.1 KQ459983 KPJ19122.1 KQ460140 KPJ17511.1 GG666659 EEN45471.1 AGBW02010415 OWR48546.1 AAWZ02035144 AAWZ02035145 AMGL01047138
Proteomes
PRIDE
Interpro
ProteinModelPortal
H9IWZ4
A0A2A4K9P1
A0A3S2NW05
A0A2A4KAB9
A0A2A4JMU5
A0A3S2THA9
+ More
A0A3S2NF42 A0A2W1BK83 A0A0N0PBL1 A0A3S2NV19 A0A2H1WUB2 A0A0N1ICJ9 A0A212FHB4 A0A2A4JGB7 A0A1E1WCP9 A0A212F827 A0A2H1W7Z1 A0A087YR65 A0A3B3UVZ4 A0A3B3YZ32 A0A0N1IEI9 A0A0N0PE54 A0A0N1IFW2 C3ZPQ0 A0A3B3UVG3 A0A212F479 A0A3Q3IRR6 L7MZU6 W5P8U9
A0A3S2NF42 A0A2W1BK83 A0A0N0PBL1 A0A3S2NV19 A0A2H1WUB2 A0A0N1ICJ9 A0A212FHB4 A0A2A4JGB7 A0A1E1WCP9 A0A212F827 A0A2H1W7Z1 A0A087YR65 A0A3B3UVZ4 A0A3B3YZ32 A0A0N1IEI9 A0A0N0PE54 A0A0N1IFW2 C3ZPQ0 A0A3B3UVG3 A0A212F479 A0A3Q3IRR6 L7MZU6 W5P8U9
PDB
5V3M
E-value=4.16372e-18,
Score=226
Ontologies
GO
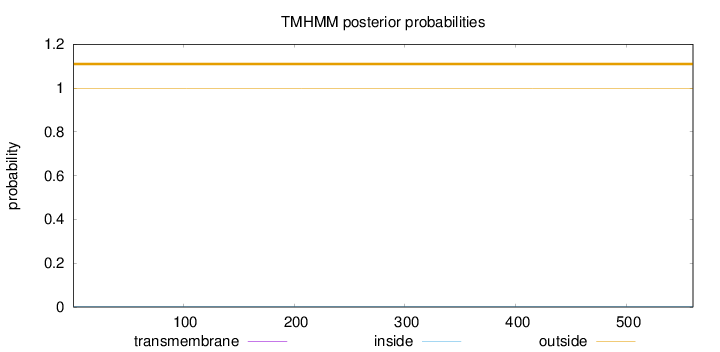
Topology
Length:
560
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00034
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00161
outside
1 - 560
Population Genetic Test Statistics
Pi
247.192644
Theta
167.326017
Tajima's D
2.225912
CLR
0.258118
CSRT
0.913054347282636
Interpretation
Uncertain
