Gene
KWMTBOMO05407
Pre Gene Modal
BGIBMGA006678
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_protein_tiptop-like_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 3.915
Sequence
CDS
ATGATTGTCGAGCCCGACCACGTGAGGGAGAGCCCGAGATGTCTTTCGCGGGAGTCGTCGGGTGCGCCGCGATGTCCCTCCAATGACTCGGTGTACTCCGGACGTAGCGGGGCTGGCCTTCCTCTGCCCGCCGCGCTCACCGCCGCTCTACCAGCCGCACTGCTTCCACCTCACTCAGCAGCGGTCGCCGCGTACCTCGGCGCAGCAGCCGCAGCCGCCGCCCAGCAACGTTTGCTCATGTCCTGCCACGAGGACCTCACTGACCCCGAACGTGCAGATGCCGTACTTGACTTTAGCACAAAACGTAGCGACTCGCCCCACGACGACGAGGATATAGAGGATGCCGTCAACCTCAGCAAAAACGAAAACGGTCCTCTGGACCTTTCGGTTGGGACGAGAAAAAGGGGACCAGAAGACTCGCCTTCACCTGTTCCCATTAGAAAAAGTTCTAGACCTGCAGAGTACAAACCAACCCCGTCTCCGTGGTCTACGCCTGTCGCCCCTCATTTACCTTATTTTGCCGCTGCAGTTGCTGCTGCCAGTCTGTCGCCTAAAGGAGGTGTTCCTGCTGACTGGAATGGTAAACTGAAGCACGGCGGCCCTACACCGAACGACGCAACTAAAGCATTAGAAAAAATGAGTGAACTTAGCAGACTCGGTGGGGAAGAACTCTTCCGATCAGTACAAACAGCTGCTTTAGGTGCCGGCTTGGCTCCGAACGCAGCCGCCAGACATTCGGCTTGGCAATCTCATTGGCTCAACAAAGGAGCGGATCAAACGAAAGATGTACTTAAATGTGTGTGGTGTAAGAAGAGCTTCAATTCTCTAGCAGATCTTACAGTTCACATGAAAGAGGCCAAGCATTGTGGTGTAAATGTGCCAGTGCCACCATCTACAGGTGCACCGATCCCACCATCCTTACAAACTCCCACTAGTTCACCATCGACTCCATCGCACAATTCATCTTCTTCTAGCGGTTCTTCGAAACCGTCACAAAGCGATTTAAATATGTTAATTAAAGAAAACATGCCGATTCCTAGAAAGCTAGTTAGGGGTCAGGACGTTTGGTTAGGTAAAGGCGCTGAGCAAACTAGGCAAATATTGAAATGTATGTGGTGTGCAGAAAGTTTTCGTTCTTTAGCTGAGATGACGAGTCACATGCAACGCACTCAGCATTATACGAATATTATTTCTCAAGAGCAAATTATTTCATGGAAATCATCTGAAGACGGAAAGGGCTCCAACGCAAGCGGTCCGGGTACTAATAATTCAGTTCCACCTACTACGGGCACCAGTAGTCACGTCAGCGCCGTTCTCACTTGTAAAGTTTGCGATCAAGCATTCAGTTCATTGAAAGAACTAAGTAATCACATGGTCAAGAACTCTCATTACAAGGAACACATAATGCGATCGATTACCGAAAGTGGAGGTAGACGACGCCAGACTCGTGAGAAGCGAAAGAAATCATTGCCAGTCAGGAAACTCCTTGAATTAGAAAGAGCCCAGCATGAATTTAAAAACGGAGACGGTAACGGCGTGCCTATGGGTAAACCAATTCGTGATTATGGCACCGGAAGCCGCATCACGTGTGAGAAATGTGGGGACAAAATCGAGACCGCAGTATTTGTGGAGCACATTAGGCAGTGTATCGGAGCACCGGTTTCTAACAGTCAAAGGAATTTTCTGAAGAGCGCATTAATGTCGAACAACATAATTCCTCCAGACGTGCCGGGACACATTACCCCCACCAGCCGTGATGGACGCAAAAGCATCAACGAAGAACTGCCTTCCCCTGGGTCCGCACACCACCGATCTCCTTCGTCGGTTAATGATTCATCTCCCTGTTCCAAAGATCCCAATGTGAACATGGACAAGAGCTCATCACCATCGGTACTTAACGCCATAGAACAGCTGATTGAGAAGAGTTTTGACACTCGATCACGACATTCAGTACCGGGAATGCCCGGAGGCGCATCTCACGCTCCGATAGGATCAAGTATCCTTAAGCGCTTGGGCATAGACGAAAGCGTTGATTATACAAAACCTTTGGTTGATGCTCAAACAATGAACATGCTCAGAAACTATCACCAGCAGTCCGGTTATGGACGGCGAGAGCGCAGCGGTAGCGAATCTAGTTCTATGTCTGAAAGAGGTGGCAGCAGGGTTGAGTCTCTCACACCCGATAGAAAGCTAGATTCATATCACATGACACCACGCACCACTCCTGATACTCGCGGCTCACAGACTCCAGCTTCCGAAGATCGACATGAAATACGGATAAAAAAAGAAGTAGACGAAGAGGATCAACGAGAAAACGGCGTAGATTTGAGTAATCAACCAGTGCGGGTGAAGACTGAGATGGAAGAAGAAGAGGAACCTCTACGACCAAGGAGCTCTACGGAAGAAGATATCAAACCAGTCGTACCCAAACGCGAGAGCGAGGGTCCCAGCCCAGCCGCTAGCCCCCGCAGTCCGTCCAGTGATCGCTCAGGCAACACTCCGGGTGCTGATAGGAAACCGGCTTCTAGTTTAGGGGCTCTTTCCTCTATGTTCGATAATCTTGCGGGAGGCGGACCGTCTAATGAGCCAAGTTCTTCCCGGCGCGGTGGCAGCCATCCTCTGGCCGCACTACAAAAGCTGTGCGACAAGACCGAGAACACACCGGCACGTGCATCTGCGCCGGCGCAATCTCCGGCCGGGCCGCCCAGCATCCTCACCTTCAGCTGGGCCTGCAACGATGCCGTCGTCACAGATTCCGTTATGAAGTGTGCGCTGTGCGATACTCCATTCATTTCGAAGGGTGCATATCGCCACCACTTATCGAAAATGCACTTTGTGAAGGACGGTGCCTTGCCGGAGCCGGTGCCTCTGAAGGCGGGGTCGCCGGCGCCGCACAAGCCTGGCTCGGCGGCTGGCTCCCCGCAGGACCCCCGCAGCCCCGCGCAGACCTTCGACGAGAGCCCGCACTCCAAGTTCCTCAAGTACACGGAACTGGCCAAGCAGCTCTCCAGCAAGTACGTGTGA
Protein
MIVEPDHVRESPRCLSRESSGAPRCPSNDSVYSGRSGAGLPLPAALTAALPAALLPPHSAAVAAYLGAAAAAAAQQRLLMSCHEDLTDPERADAVLDFSTKRSDSPHDDEDIEDAVNLSKNENGPLDLSVGTRKRGPEDSPSPVPIRKSSRPAEYKPTPSPWSTPVAPHLPYFAAAVAAASLSPKGGVPADWNGKLKHGGPTPNDATKALEKMSELSRLGGEELFRSVQTAALGAGLAPNAAARHSAWQSHWLNKGADQTKDVLKCVWCKKSFNSLADLTVHMKEAKHCGVNVPVPPSTGAPIPPSLQTPTSSPSTPSHNSSSSSGSSKPSQSDLNMLIKENMPIPRKLVRGQDVWLGKGAEQTRQILKCMWCAESFRSLAEMTSHMQRTQHYTNIISQEQIISWKSSEDGKGSNASGPGTNNSVPPTTGTSSHVSAVLTCKVCDQAFSSLKELSNHMVKNSHYKEHIMRSITESGGRRRQTREKRKKSLPVRKLLELERAQHEFKNGDGNGVPMGKPIRDYGTGSRITCEKCGDKIETAVFVEHIRQCIGAPVSNSQRNFLKSALMSNNIIPPDVPGHITPTSRDGRKSINEELPSPGSAHHRSPSSVNDSSPCSKDPNVNMDKSSSPSVLNAIEQLIEKSFDTRSRHSVPGMPGGASHAPIGSSILKRLGIDESVDYTKPLVDAQTMNMLRNYHQQSGYGRRERSGSESSSMSERGGSRVESLTPDRKLDSYHMTPRTTPDTRGSQTPASEDRHEIRIKKEVDEEDQRENGVDLSNQPVRVKTEMEEEEEPLRPRSSTEEDIKPVVPKRESEGPSPAASPRSPSSDRSGNTPGADRKPASSLGALSSMFDNLAGGGPSNEPSSSRRGGSHPLAALQKLCDKTENTPARASAPAQSPAGPPSILTFSWACNDAVVTDSVMKCALCDTPFISKGAYRHHLSKMHFVKDGALPEPVPLKAGSPAPHKPGSAAGSPQDPRSPAQTFDESPHSKFLKYTELAKQLSSKYV
Summary
Uniprot
A0A1C9EGJ1
A0A212EVI1
A0A2H1WUG9
A0A194QHE5
A0A0N1I799
A0A0L7LKY3
+ More
H9JAY3 A0A1E1WA08 D6X1P2 Q967X9 A0A0T6BCK3 A0A1B0CDQ8 A0A1W4WVG6 T1HHQ8 A0A1B0DRC9 A0A336LKI7 E0V8X9 A0A1B0GL62 A0A3Q0JR35 Q7PWB2 A0A182RNF9 A0A182H315 A0A1S4FHB9 A0A1Y1KB80 A0A182QPF9 A0A2P8YHP1 A0A2J7QDP7 A0A067QU69 A0A023F1Q3 A0A2R7WTX0 A0A0A9XIX9 A0A182K6Z6 A0A0L0BND2 A0A1A9WXB3 K7J8N6 A0A182TC34 A0A1I8MCW0 A0A182NBD3 A0A084VKM2 A0A182PNG9 B4GKM3 A0A0P8XHN0 B5DHA1 A0A3B0KA05 W8BZS7 A0A1B0BBR3 A0A1A9XLF6 A0A1I8PTG1 B4III1 B4P6F0 A0A182W9Z6 A0A182M973
H9JAY3 A0A1E1WA08 D6X1P2 Q967X9 A0A0T6BCK3 A0A1B0CDQ8 A0A1W4WVG6 T1HHQ8 A0A1B0DRC9 A0A336LKI7 E0V8X9 A0A1B0GL62 A0A3Q0JR35 Q7PWB2 A0A182RNF9 A0A182H315 A0A1S4FHB9 A0A1Y1KB80 A0A182QPF9 A0A2P8YHP1 A0A2J7QDP7 A0A067QU69 A0A023F1Q3 A0A2R7WTX0 A0A0A9XIX9 A0A182K6Z6 A0A0L0BND2 A0A1A9WXB3 K7J8N6 A0A182TC34 A0A1I8MCW0 A0A182NBD3 A0A084VKM2 A0A182PNG9 B4GKM3 A0A0P8XHN0 B5DHA1 A0A3B0KA05 W8BZS7 A0A1B0BBR3 A0A1A9XLF6 A0A1I8PTG1 B4III1 B4P6F0 A0A182W9Z6 A0A182M973
Pubmed
EMBL
KX778609
AON96607.1
AGBW02012180
OWR45502.1
ODYU01011150
SOQ56715.1
+ More
KQ458793 KPJ04968.1 KQ461033 KPJ09967.1 JTDY01000726 KOB76090.1 BABH01008287 GDQN01007214 JAT83840.1 KQ971371 EFA10732.1 AF356647 AAK43700.1 LJIG01001884 KRT85056.1 AJWK01008139 AJWK01008140 AJWK01008141 AJWK01008142 ACPB03000468 AJVK01009039 AJVK01009040 AJVK01009041 UFQS01000003 UFQT01000003 SSW96736.1 SSX17123.1 DS234986 EEB09835.1 AJWK01034351 AAAB01008984 EAA14779.5 JXUM01106407 KQ565052 KXJ71407.1 GEZM01087322 JAV58772.1 AXCN02000626 PYGN01000587 PSN43770.1 NEVH01015817 PNF26705.1 KK852946 KDR13419.1 GBBI01003469 JAC15243.1 KK855497 PTY22831.1 GBHO01023655 JAG19949.1 JRES01001605 KNC21537.1 AAZX01000111 AAZX01000114 ATLV01014233 KE524948 KFB38516.1 CH479184 EDW37189.1 CH902624 KPU74390.1 CH379058 EDY69676.1 OUUW01000022 SPP89542.1 GAMC01004332 JAC02224.1 JXJN01011612 CH480843 EDW49731.1 CM000158 EDW89907.1 AXCM01001343
KQ458793 KPJ04968.1 KQ461033 KPJ09967.1 JTDY01000726 KOB76090.1 BABH01008287 GDQN01007214 JAT83840.1 KQ971371 EFA10732.1 AF356647 AAK43700.1 LJIG01001884 KRT85056.1 AJWK01008139 AJWK01008140 AJWK01008141 AJWK01008142 ACPB03000468 AJVK01009039 AJVK01009040 AJVK01009041 UFQS01000003 UFQT01000003 SSW96736.1 SSX17123.1 DS234986 EEB09835.1 AJWK01034351 AAAB01008984 EAA14779.5 JXUM01106407 KQ565052 KXJ71407.1 GEZM01087322 JAV58772.1 AXCN02000626 PYGN01000587 PSN43770.1 NEVH01015817 PNF26705.1 KK852946 KDR13419.1 GBBI01003469 JAC15243.1 KK855497 PTY22831.1 GBHO01023655 JAG19949.1 JRES01001605 KNC21537.1 AAZX01000111 AAZX01000114 ATLV01014233 KE524948 KFB38516.1 CH479184 EDW37189.1 CH902624 KPU74390.1 CH379058 EDY69676.1 OUUW01000022 SPP89542.1 GAMC01004332 JAC02224.1 JXJN01011612 CH480843 EDW49731.1 CM000158 EDW89907.1 AXCM01001343
Proteomes
UP000007151
UP000053268
UP000053240
UP000037510
UP000005204
UP000007266
+ More
UP000092461 UP000192223 UP000015103 UP000092462 UP000009046 UP000079169 UP000007062 UP000075900 UP000069940 UP000249989 UP000075886 UP000245037 UP000235965 UP000027135 UP000075881 UP000037069 UP000091820 UP000002358 UP000075901 UP000095301 UP000075884 UP000030765 UP000075885 UP000008744 UP000007801 UP000001819 UP000268350 UP000092460 UP000092443 UP000095300 UP000001292 UP000002282 UP000075920 UP000075883
UP000092461 UP000192223 UP000015103 UP000092462 UP000009046 UP000079169 UP000007062 UP000075900 UP000069940 UP000249989 UP000075886 UP000245037 UP000235965 UP000027135 UP000075881 UP000037069 UP000091820 UP000002358 UP000075901 UP000095301 UP000075884 UP000030765 UP000075885 UP000008744 UP000007801 UP000001819 UP000268350 UP000092460 UP000092443 UP000095300 UP000001292 UP000002282 UP000075920 UP000075883
PRIDE
Interpro
SUPFAM
SSF57667
SSF57667
ProteinModelPortal
A0A1C9EGJ1
A0A212EVI1
A0A2H1WUG9
A0A194QHE5
A0A0N1I799
A0A0L7LKY3
+ More
H9JAY3 A0A1E1WA08 D6X1P2 Q967X9 A0A0T6BCK3 A0A1B0CDQ8 A0A1W4WVG6 T1HHQ8 A0A1B0DRC9 A0A336LKI7 E0V8X9 A0A1B0GL62 A0A3Q0JR35 Q7PWB2 A0A182RNF9 A0A182H315 A0A1S4FHB9 A0A1Y1KB80 A0A182QPF9 A0A2P8YHP1 A0A2J7QDP7 A0A067QU69 A0A023F1Q3 A0A2R7WTX0 A0A0A9XIX9 A0A182K6Z6 A0A0L0BND2 A0A1A9WXB3 K7J8N6 A0A182TC34 A0A1I8MCW0 A0A182NBD3 A0A084VKM2 A0A182PNG9 B4GKM3 A0A0P8XHN0 B5DHA1 A0A3B0KA05 W8BZS7 A0A1B0BBR3 A0A1A9XLF6 A0A1I8PTG1 B4III1 B4P6F0 A0A182W9Z6 A0A182M973
H9JAY3 A0A1E1WA08 D6X1P2 Q967X9 A0A0T6BCK3 A0A1B0CDQ8 A0A1W4WVG6 T1HHQ8 A0A1B0DRC9 A0A336LKI7 E0V8X9 A0A1B0GL62 A0A3Q0JR35 Q7PWB2 A0A182RNF9 A0A182H315 A0A1S4FHB9 A0A1Y1KB80 A0A182QPF9 A0A2P8YHP1 A0A2J7QDP7 A0A067QU69 A0A023F1Q3 A0A2R7WTX0 A0A0A9XIX9 A0A182K6Z6 A0A0L0BND2 A0A1A9WXB3 K7J8N6 A0A182TC34 A0A1I8MCW0 A0A182NBD3 A0A084VKM2 A0A182PNG9 B4GKM3 A0A0P8XHN0 B5DHA1 A0A3B0KA05 W8BZS7 A0A1B0BBR3 A0A1A9XLF6 A0A1I8PTG1 B4III1 B4P6F0 A0A182W9Z6 A0A182M973
PDB
2DMI
E-value=0.0011377,
Score=104
Ontologies
GO
PANTHER
Topology
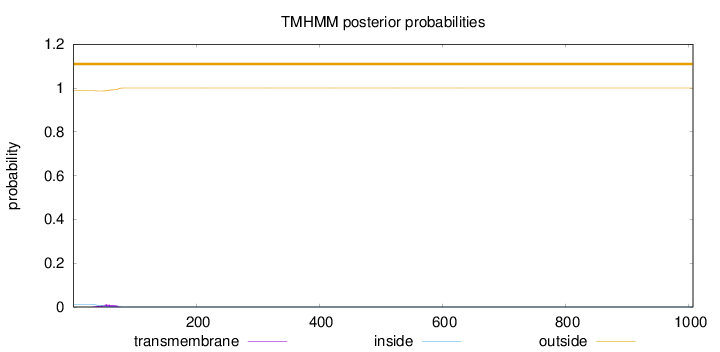
Length:
1007
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.29547
Exp number, first 60 AAs:
0.18647
Total prob of N-in:
0.01276
outside
1 - 1007
Population Genetic Test Statistics
Pi
207.68357
Theta
188.452591
Tajima's D
0.413387
CLR
0.531608
CSRT
0.488025598720064
Interpretation
Uncertain
