Gene
KWMTBOMO04908
Pre Gene Modal
BGIBMGA008014
Annotation
PREDICTED:_uncharacterized_protein_LOC101744688_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 2.335
Sequence
CDS
ATGATCGCTAATCTGCCGCTGTTGTTCGCCAATGGCAATCCGATACCGCTTTCGAAGATCAGAGCCGGCGAGCTCGAGAGGTTTATAACATTTATGGTGACATGTTCCTGGGGTCACGATACAGCTAAGGATATTCGACAGCCTCCATGGTGGCCCAGCAACGTCGAGTTCACGCATCCCTTCGTGAGGCCTGCAGCTGTGCCCAACGATTGGGAGCTCAGGTTGAAAAATGTTGTCAAAAAATGCTACGAATACCACAAGAGTGCATTTCTGCTGGTCTTTTCAGCGCAACTGTCGCGATACCCCCGTAACCGTCTTTTGTACGTTGACAACAAGGATCACACAACATCCCTTTATTATATACCTAGTAACCGTCTATTGGTTACATTCAGAAATGAGAATATAAACTATGATAAGGATGTACAAATTGAAGACGAAGAGCCTGTGGTGAGAGCTGCAGACATTTATTTGTGTGACAATTGTGATAGTCATTTTGATAATTTAAACAATTTACAAGTCCACGAGAGACTGTGTCATGGTGACAACTCTCCTGCTGGCACGAGTGGTGGTTTACCAGAGTTCTTAGCTGCATTGAAGTTAAAACAGGCTACAGATGTTCAATGCTCAGGGTCTGAACATATAGAATATGAAGCAAGACCTAAGAACGCTAGGAATGCTGCAAATATTGACCGTGGGCCACCTTATCCCTTTTCATCATTAGCTTACATTAAAAATGCTAAAACATTAGTACAGAGAGATACAAGTTATTCAAGGGAGAGAATTGAACGCTACTGTTGTACTTCCACTCCCCTTAGTAAAAATGTAGTTAGTAAAAATAAACATCAACAATTCCCTGTAAGATATAAGAGGCCTATAGATTATTGGCACAGAAAGCATGTGTTCCCTGACCAGAGAAATAAAGATATATTAGATCTTAAAGCTCAATTGTGTTTATTGAAATGCAGAGTCCTCACTGTTAATGTTGAAAGAATGTCTGAAGAAAAAATAGAAGACTATATTCAACAGTGCAAAAATAAATTGGAAAGTGAACTACAACAGAAACAAGAAAAAGATGTTATATTTGTGGACAATATAGACACTGATCAAGGAGAAGATGAAGAAACAAAGATTGCAGACCCGTTGAGAAGATCTGAAGTAGAATGTGAAGTGATTGACCTTTGTTCTGATGATGAGAGCAGTAGTAGTAATGAGAATTGTGACCCACGTGCAGGTGTGGCTTGTGTGATGAGAGGTGGAGCAATTTTGAGACGAGCAGCTGCATCGACACCACAAGCACTCCCCGCTGATCCATGTGGCGCCAGGCATCTCCCAGCAGTACTGCAACCTCACCCCAATCCTCAACCTATCATACTCATAGCCCATACATTTAATAATCTACAGACTATATCTTTAGAGTAA
Protein
MIANLPLLFANGNPIPLSKIRAGELERFITFMVTCSWGHDTAKDIRQPPWWPSNVEFTHPFVRPAAVPNDWELRLKNVVKKCYEYHKSAFLLVFSAQLSRYPRNRLLYVDNKDHTTSLYYIPSNRLLVTFRNENINYDKDVQIEDEEPVVRAADIYLCDNCDSHFDNLNNLQVHERLCHGDNSPAGTSGGLPEFLAALKLKQATDVQCSGSEHIEYEARPKNARNAANIDRGPPYPFSSLAYIKNAKTLVQRDTSYSRERIERYCCTSTPLSKNVVSKNKHQQFPVRYKRPIDYWHRKHVFPDQRNKDILDLKAQLCLLKCRVLTVNVERMSEEKIEDYIQQCKNKLESELQQKQEKDVIFVDNIDTDQGEDEETKIADPLRRSEVECEVIDLCSDDESSSSNENCDPRAGVACVMRGGAILRRAAASTPQALPADPCGARHLPAVLQPHPNPQPIILIAHTFNNLQTISLE
Summary
Uniprot
H9JER7
A0A194QLX7
A0A194QCU5
A0A2A4JGK4
A0A2W1BKU2
A0A212F6M3
+ More
A0A2H1WZV5 A0A1E1WFX1 A0A0L7LQT1 S4PM39 A0A151IDY4 A0A088AUW6 A0A2A3E7B1 E2BTC5 E9J5F9 A0A154PIP6 A0A026WQR5 A0A151K144 A0A158NV76 A0A151JCB0 A0A310SI81 A0A195B639 F4WPY0 K7JEX9 A0A0C9R0L8 A0A0C9PHP1 E2ANP0 A0A3L8DQB7 A0A151X2H5 A0A0L7QLG1 A0A336M0Y2 A0A336LT97 A0A0K8W1X8 A0A1I8QCZ2 A0A1B0FR23 A0A1A9UCR7 A0A1B0CHR2 A0A1I8ME92 A0A1A9WEX8 Q9VL20 A0A0A1XKX7 A0A182P4E7 A0A1W4UFH5 A0A1L8DB62 B4Q8R1 B3N942 A0A0Q5WJ91 W8BY72 B4HWH8 A0A2J7PTM1 A0A2J7PTL3 A0A2J7PTL0 A0A2J7PTL9 A0A023ES38 A0A2J7PTM5 A0A0P6IUH9 A0A0R1DKY1 B4NZ66 B4JPN4 B4KHP2 A0A0Q9XGI7 A0A3B0JGS0 B3MPN2 A0A067R4I3 A0A1J1HTD6 E9FVF5
A0A2H1WZV5 A0A1E1WFX1 A0A0L7LQT1 S4PM39 A0A151IDY4 A0A088AUW6 A0A2A3E7B1 E2BTC5 E9J5F9 A0A154PIP6 A0A026WQR5 A0A151K144 A0A158NV76 A0A151JCB0 A0A310SI81 A0A195B639 F4WPY0 K7JEX9 A0A0C9R0L8 A0A0C9PHP1 E2ANP0 A0A3L8DQB7 A0A151X2H5 A0A0L7QLG1 A0A336M0Y2 A0A336LT97 A0A0K8W1X8 A0A1I8QCZ2 A0A1B0FR23 A0A1A9UCR7 A0A1B0CHR2 A0A1I8ME92 A0A1A9WEX8 Q9VL20 A0A0A1XKX7 A0A182P4E7 A0A1W4UFH5 A0A1L8DB62 B4Q8R1 B3N942 A0A0Q5WJ91 W8BY72 B4HWH8 A0A2J7PTM1 A0A2J7PTL3 A0A2J7PTL0 A0A2J7PTL9 A0A023ES38 A0A2J7PTM5 A0A0P6IUH9 A0A0R1DKY1 B4NZ66 B4JPN4 B4KHP2 A0A0Q9XGI7 A0A3B0JGS0 B3MPN2 A0A067R4I3 A0A1J1HTD6 E9FVF5
Pubmed
19121390
26354079
28756777
22118469
26227816
23622113
+ More
20798317 21282665 24508170 21347285 21719571 20075255 30249741 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 17994087 22936249 18057021 24495485 24945155 26999592 17550304 24845553 21292972
20798317 21282665 24508170 21347285 21719571 20075255 30249741 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 17994087 22936249 18057021 24495485 24945155 26999592 17550304 24845553 21292972
EMBL
BABH01023421
KQ461198
KPJ05965.1
KQ459185
KPJ03363.1
NWSH01001607
+ More
PCG70714.1 KZ150199 PZC72293.1 AGBW02009996 OWR49374.1 ODYU01012276 SOQ58496.1 GDQN01005171 JAT85883.1 JTDY01000358 KOB77556.1 GAIX01000311 JAA92249.1 KQ977901 KYM98896.1 KZ288356 PBC27172.1 GL450389 EFN81046.1 GL768153 EFZ11945.1 KQ434926 KZC11687.1 KK107128 EZA58387.1 KQ981189 KYN45249.1 ADTU01026946 KQ979060 KYN23170.1 KQ759866 OAD62488.1 KQ976595 KYM79644.1 GL888256 EGI63748.1 AAZX01003294 AAZX01005611 GBYB01000386 JAG70153.1 GBYB01000388 JAG70155.1 GL441283 EFN64951.1 QOIP01000006 RLU21998.1 KQ982577 KYQ54637.1 KQ414934 KOC59346.1 UFQT01000093 SSX19628.1 SSX19627.1 GDHF01007177 JAI45137.1 CCAG010018514 AJWK01012600 AE014134 BT001618 AAF52878.2 AAN71373.1 GBXI01002646 JAD11646.1 GFDF01010386 JAV03698.1 CM000361 CM002910 EDX04476.1 KMY89435.1 CH954177 EDV58477.2 KQS70412.1 KQS70413.1 KQS70414.1 GAMC01012356 JAB94199.1 CH480818 EDW52373.1 NEVH01021236 PNF19675.1 PNF19673.1 PNF19676.1 PNF19671.1 GAPW01001548 JAC12050.1 PNF19674.1 GDUN01000503 JAN95416.1 CM000157 KRJ98011.1 EDW88761.1 KRJ98008.1 KRJ98009.1 KRJ98010.1 KRJ98012.1 CH916372 EDV98864.1 CH933807 EDW12321.2 KRG03190.1 KRG03191.1 OUUW01000006 SPP81584.1 CH902620 EDV32280.1 KPU73878.1 KK852701 KDR18139.1 CVRI01000020 CRK90644.1 GL732525 EFX89113.1
PCG70714.1 KZ150199 PZC72293.1 AGBW02009996 OWR49374.1 ODYU01012276 SOQ58496.1 GDQN01005171 JAT85883.1 JTDY01000358 KOB77556.1 GAIX01000311 JAA92249.1 KQ977901 KYM98896.1 KZ288356 PBC27172.1 GL450389 EFN81046.1 GL768153 EFZ11945.1 KQ434926 KZC11687.1 KK107128 EZA58387.1 KQ981189 KYN45249.1 ADTU01026946 KQ979060 KYN23170.1 KQ759866 OAD62488.1 KQ976595 KYM79644.1 GL888256 EGI63748.1 AAZX01003294 AAZX01005611 GBYB01000386 JAG70153.1 GBYB01000388 JAG70155.1 GL441283 EFN64951.1 QOIP01000006 RLU21998.1 KQ982577 KYQ54637.1 KQ414934 KOC59346.1 UFQT01000093 SSX19628.1 SSX19627.1 GDHF01007177 JAI45137.1 CCAG010018514 AJWK01012600 AE014134 BT001618 AAF52878.2 AAN71373.1 GBXI01002646 JAD11646.1 GFDF01010386 JAV03698.1 CM000361 CM002910 EDX04476.1 KMY89435.1 CH954177 EDV58477.2 KQS70412.1 KQS70413.1 KQS70414.1 GAMC01012356 JAB94199.1 CH480818 EDW52373.1 NEVH01021236 PNF19675.1 PNF19673.1 PNF19676.1 PNF19671.1 GAPW01001548 JAC12050.1 PNF19674.1 GDUN01000503 JAN95416.1 CM000157 KRJ98011.1 EDW88761.1 KRJ98008.1 KRJ98009.1 KRJ98010.1 KRJ98012.1 CH916372 EDV98864.1 CH933807 EDW12321.2 KRG03190.1 KRG03191.1 OUUW01000006 SPP81584.1 CH902620 EDV32280.1 KPU73878.1 KK852701 KDR18139.1 CVRI01000020 CRK90644.1 GL732525 EFX89113.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000007151
UP000037510
+ More
UP000078542 UP000005203 UP000242457 UP000008237 UP000076502 UP000053097 UP000078541 UP000005205 UP000078492 UP000078540 UP000007755 UP000002358 UP000000311 UP000279307 UP000075809 UP000053825 UP000095300 UP000092444 UP000078200 UP000092461 UP000095301 UP000091820 UP000000803 UP000075885 UP000192221 UP000000304 UP000008711 UP000001292 UP000235965 UP000002282 UP000001070 UP000009192 UP000268350 UP000007801 UP000027135 UP000183832 UP000000305
UP000078542 UP000005203 UP000242457 UP000008237 UP000076502 UP000053097 UP000078541 UP000005205 UP000078492 UP000078540 UP000007755 UP000002358 UP000000311 UP000279307 UP000075809 UP000053825 UP000095300 UP000092444 UP000078200 UP000092461 UP000095301 UP000091820 UP000000803 UP000075885 UP000192221 UP000000304 UP000008711 UP000001292 UP000235965 UP000002282 UP000001070 UP000009192 UP000268350 UP000007801 UP000027135 UP000183832 UP000000305
PRIDE
Pfam
PF10491 Nrf1_DNA-bind
ProteinModelPortal
H9JER7
A0A194QLX7
A0A194QCU5
A0A2A4JGK4
A0A2W1BKU2
A0A212F6M3
+ More
A0A2H1WZV5 A0A1E1WFX1 A0A0L7LQT1 S4PM39 A0A151IDY4 A0A088AUW6 A0A2A3E7B1 E2BTC5 E9J5F9 A0A154PIP6 A0A026WQR5 A0A151K144 A0A158NV76 A0A151JCB0 A0A310SI81 A0A195B639 F4WPY0 K7JEX9 A0A0C9R0L8 A0A0C9PHP1 E2ANP0 A0A3L8DQB7 A0A151X2H5 A0A0L7QLG1 A0A336M0Y2 A0A336LT97 A0A0K8W1X8 A0A1I8QCZ2 A0A1B0FR23 A0A1A9UCR7 A0A1B0CHR2 A0A1I8ME92 A0A1A9WEX8 Q9VL20 A0A0A1XKX7 A0A182P4E7 A0A1W4UFH5 A0A1L8DB62 B4Q8R1 B3N942 A0A0Q5WJ91 W8BY72 B4HWH8 A0A2J7PTM1 A0A2J7PTL3 A0A2J7PTL0 A0A2J7PTL9 A0A023ES38 A0A2J7PTM5 A0A0P6IUH9 A0A0R1DKY1 B4NZ66 B4JPN4 B4KHP2 A0A0Q9XGI7 A0A3B0JGS0 B3MPN2 A0A067R4I3 A0A1J1HTD6 E9FVF5
A0A2H1WZV5 A0A1E1WFX1 A0A0L7LQT1 S4PM39 A0A151IDY4 A0A088AUW6 A0A2A3E7B1 E2BTC5 E9J5F9 A0A154PIP6 A0A026WQR5 A0A151K144 A0A158NV76 A0A151JCB0 A0A310SI81 A0A195B639 F4WPY0 K7JEX9 A0A0C9R0L8 A0A0C9PHP1 E2ANP0 A0A3L8DQB7 A0A151X2H5 A0A0L7QLG1 A0A336M0Y2 A0A336LT97 A0A0K8W1X8 A0A1I8QCZ2 A0A1B0FR23 A0A1A9UCR7 A0A1B0CHR2 A0A1I8ME92 A0A1A9WEX8 Q9VL20 A0A0A1XKX7 A0A182P4E7 A0A1W4UFH5 A0A1L8DB62 B4Q8R1 B3N942 A0A0Q5WJ91 W8BY72 B4HWH8 A0A2J7PTM1 A0A2J7PTL3 A0A2J7PTL0 A0A2J7PTL9 A0A023ES38 A0A2J7PTM5 A0A0P6IUH9 A0A0R1DKY1 B4NZ66 B4JPN4 B4KHP2 A0A0Q9XGI7 A0A3B0JGS0 B3MPN2 A0A067R4I3 A0A1J1HTD6 E9FVF5
Ontologies
GO
PANTHER
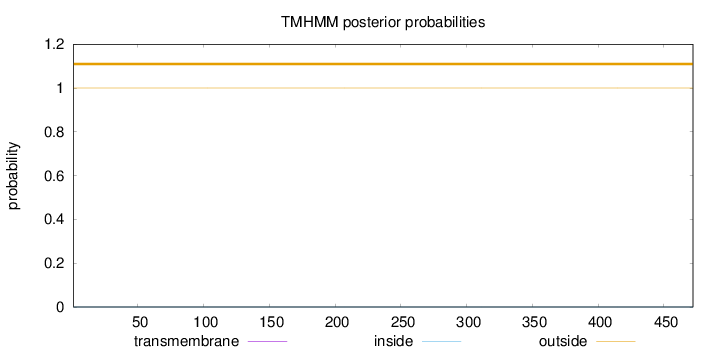
Topology
Length:
472
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000920000000000001
Exp number, first 60 AAs:
0.00038
Total prob of N-in:
0.00032
outside
1 - 472
Population Genetic Test Statistics
Pi
153.980557
Theta
205.662142
Tajima's D
-0.528196
CLR
0.722854
CSRT
0.23768811559422
Interpretation
Uncertain
