Gene
KWMTBOMO04442
Pre Gene Modal
BGIBMGA005416
Annotation
methoprene-tolerant_protein_1_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 4.138
Sequence
CDS
ATGAACTCGTCTATTGGGACAGGATCGAATTACGAGAAATGCTCTAAGATGGCAACTCGGGAGCCGGCAGAGTTTCTGCCCGTGGCAGTGACAACAAATTGCGTTTACTATGATTCACAAGCTGCAAGCTGCGGAGACTCTTCTTCAGATTCACGTTCGCCAGAGACTACTTCAGCAAAGGCTAGCGATCGTGCTTCTCGAATCATTGCAGAAAAAACTCGTCGCAGCCAGTACAATGCATTAATCCATCAAATGAAGTCTCTCCTATCGGACATTGCGCATTCCCAACGGAAAGTTGACAAAACTAGTATTTTGAGACACGCCGTCAACAAACTGCGTAACGAACACGTCTTTGGCGATACCATTAAATGCTGCCATCTAGAAACATGGTCCCCTGCATTTTTAAAGTTTTTTGATCTCATAGGAGGTATCATGTTTGCTGTCACTTGTCGGGGTCGCATTTTTATAGTTTCACCAAATATCCAAGAAAAATTAGGTTATTGCCATATAGATTTAGTAGGCCAGGATTTTTACAAATATGTTCATGAGGAAGACAGAGAAACCCTACGTTGTCACATTTACCCACCTGAACTTCAAACAGGCTGTGATGATCAACTTTTTGAACATAACCATAACTTTCATATTCGTCTGATGAGAGCAGGAGCAAGGTCTGATCATCCCAGATATGAAAAATGTCGTATTAGTGGTATGTTAAGAAAGTCTGATAGAGCTACTGGGAATGAAGTTCAAGATGAGCATGTAGTTAGAAGGCAAAGAGTACGGAATAATCGCACATTTTCCTCCAGTGGAAATGACTTTGTTTTTATTGGTATGATACGTGTGCTTTCTAGCAACTTGCCAGCTCGAATCTTGCCACCAACAGCTTATTCTGAATATTGGACGAGACATTTGATAGATGGCCGGATAGTACAATGTGATCAAAGCATATCATTGGCAGCTGGTTACATGACTGATGAAGTAACAGGTACTTCAGCTTTTGTTTTTATGCACAAAGAAGATGTGCGTTGGGTTATCTGTGTACTTCGTCAAATGTATGATCAGAGTCGAGAGTTTGGGGAATCATATTACAGGTTAATATCTCGTTCTGGTCACTTCATATATATGAGAACTAAAGGATATCTTGAAATTGATAAAAAAACTAAAAAAGTTCAAAGTTTTGTCTGTGTTAATACTGTTATTGGTGAGGAGTTTGGAAAAAGAATGATGGAAGAAATGAAACGAAAGTACTCAGTTATCGTAGGAATGGATAAACAACAACAAGAAAGAGTGTTGACTTATGATGATGCTCCGGTTGAACATCCAAAGTGTCTTGAGCGGATTGTAATGCATCTTGTGGAACCTTCCGTAAGTGAAAATGTAGATGAACTGAAGTTGGTGCCTCCTTCTAGAGAGAACATAATTTCTGCTATCAAAAGCAGTGAGAAAGTGGTTCAAGAAACTGGTATACGTTTTGAATCAAGAAAAAGGAAAAAAAATAATATAGATGACAGCTATGAGTATTTAAAAAGGCACAGTGGATTGTAA
Protein
MNSSIGTGSNYEKCSKMATREPAEFLPVAVTTNCVYYDSQAASCGDSSSDSRSPETTSAKASDRASRIIAEKTRRSQYNALIHQMKSLLSDIAHSQRKVDKTSILRHAVNKLRNEHVFGDTIKCCHLETWSPAFLKFFDLIGGIMFAVTCRGRIFIVSPNIQEKLGYCHIDLVGQDFYKYVHEEDRETLRCHIYPPELQTGCDDQLFEHNHNFHIRLMRAGARSDHPRYEKCRISGMLRKSDRATGNEVQDEHVVRRQRVRNNRTFSSSGNDFVFIGMIRVLSSNLPARILPPTAYSEYWTRHLIDGRIVQCDQSISLAAGYMTDEVTGTSAFVFMHKEDVRWVICVLRQMYDQSREFGESYYRLISRSGHFIYMRTKGYLEIDKKTKKVQSFVCVNTVIGEEFGKRMMEEMKRKYSVIVGMDKQQQERVLTYDDAPVEHPKCLERIVMHLVEPSVSENVDELKLVPPSRENIISAIKSSEKVVQETGIRFESRKRKKNNIDDSYEYLKRHSGL
Summary
Uniprot
B0LL81
H9J7C4
B6VB33
A0A3Q8UB92
A0A023PPA4
A0A1E1WMG6
+ More
A0A2H1WH64 A0A0L7LG82 A0A1B2INL9 A0A290DSG2 A0A212FEP1 A0A194RBM2 U5ER17 A0A1L8DG79 A0A336KCZ4 B4R561 A0A336LQY1 A0A1L8DHG9 A0A1L8DHK9 A0A1L8DHA0 A0A182RPF2 A0A2H1VXM9 A0A182MNT8 A0A182KPF2 Q29JD8 Q9VXW7 A0A182PJJ1 B4PWL1 A0A182YCW7 A0A1B0CQB9 B3NTM6 A0A182NKC9 A0A182WKJ3 A0A182QYH5 Q5EFD7 A0A182ISC3 A0A1Q3FTU3 A0A182FCQ9 Q5EE11 Q17KB5 Q58FS0 Q4ZH01 A0A182GI95 B3MR82 A0A084W0Q6 A0A1Q3FTH3 B4ILF8 A0A2M3YZM3 A0A2M4BDB1 A0A1Q3FTK2 A0A1J1I5W3 A0A1S4EZU7 A0A2K9P5C3 Q7Q663 A0A3B0J9S9 T1PAE6 A0A0L0BLM0 B4JX85 A0A2M4A0I1 A0A2M4A0G3 Q2PT35 B4N211 B4M788 A0A2W1BSZ9 A0A1W4VFK7 A0A2M4BAV8 A0A1I8NGB4 A0A0M3QZ86 A0A1I8PHG3 B4L5Q0 A0A2M4CZ07 A0A1A9WWZ6 A0A1A9WAP8 A0A1B0ALM4 A0A1B0ABZ5 R4IJP6 A0A1A9ULG0 W5JN31 A0A1A9X7T0 A0A2M4CXT4 A0A182I1Q7 R4IJL6 A0A212FDU0 A0A1A9ZQE4 A0A0A1XMM1 A0A1B0BDG6 A0A1A9YJ64 D4Q9H9 B0LL82 A0A1A9UE24 A0A182X776 A0A0K8VAK9 A0A3G2KQA7 A0A034W909 A0A182HFD7 A0A0A1XNH9 W8B2P5
A0A2H1WH64 A0A0L7LG82 A0A1B2INL9 A0A290DSG2 A0A212FEP1 A0A194RBM2 U5ER17 A0A1L8DG79 A0A336KCZ4 B4R561 A0A336LQY1 A0A1L8DHG9 A0A1L8DHK9 A0A1L8DHA0 A0A182RPF2 A0A2H1VXM9 A0A182MNT8 A0A182KPF2 Q29JD8 Q9VXW7 A0A182PJJ1 B4PWL1 A0A182YCW7 A0A1B0CQB9 B3NTM6 A0A182NKC9 A0A182WKJ3 A0A182QYH5 Q5EFD7 A0A182ISC3 A0A1Q3FTU3 A0A182FCQ9 Q5EE11 Q17KB5 Q58FS0 Q4ZH01 A0A182GI95 B3MR82 A0A084W0Q6 A0A1Q3FTH3 B4ILF8 A0A2M3YZM3 A0A2M4BDB1 A0A1Q3FTK2 A0A1J1I5W3 A0A1S4EZU7 A0A2K9P5C3 Q7Q663 A0A3B0J9S9 T1PAE6 A0A0L0BLM0 B4JX85 A0A2M4A0I1 A0A2M4A0G3 Q2PT35 B4N211 B4M788 A0A2W1BSZ9 A0A1W4VFK7 A0A2M4BAV8 A0A1I8NGB4 A0A0M3QZ86 A0A1I8PHG3 B4L5Q0 A0A2M4CZ07 A0A1A9WWZ6 A0A1A9WAP8 A0A1B0ALM4 A0A1B0ABZ5 R4IJP6 A0A1A9ULG0 W5JN31 A0A1A9X7T0 A0A2M4CXT4 A0A182I1Q7 R4IJL6 A0A212FDU0 A0A1A9ZQE4 A0A0A1XMM1 A0A1B0BDG6 A0A1A9YJ64 D4Q9H9 B0LL82 A0A1A9UE24 A0A182X776 A0A0K8VAK9 A0A3G2KQA7 A0A034W909 A0A182HFD7 A0A0A1XNH9 W8B2P5
Pubmed
23300902
22753472
19121390
26227816
22118469
26354079
+ More
17994087 20966253 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25244985 17166512 17510324 26483478 24438588 12364791 14747013 17210077 26108605 28756777 25315136 23499946 20920257 23761445 25830018 25348373 24495485
17994087 20966253 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25244985 17166512 17510324 26483478 24438588 12364791 14747013 17210077 26108605 28756777 25315136 23499946 20920257 23761445 25830018 25348373 24495485
EMBL
EU249371
AB359911
ABY76058.1
BAJ05085.1
BABH01013637
FJ361198
+ More
ACJ04052.1 MH050927 AZL94115.1 KJ184572 KJ825895 AHX26585.1 AJW29006.1 GDQN01002916 JAT88138.1 ODYU01008646 SOQ52409.1 JTDY01001246 KOB74415.1 KT361877 ANZ54967.1 KX821731 ATB19377.1 AGBW02008921 OWR52183.1 KQ460397 KPJ15233.1 GANO01003978 JAB55893.1 GFDF01008622 JAV05462.1 UFQS01000067 UFQT01000067 SSW98950.1 SSX19332.1 CM000366 EDX17962.1 SSW98949.1 SSX19331.1 GFDF01008259 JAV05825.1 GFDF01008260 JAV05824.1 GFDF01008261 JAV05823.1 ODYU01005061 SOQ45568.1 AXCM01004842 CH379063 EAL32363.3 AE014298 BT132669 AAF48439.2 ADV37675.1 AEQ28953.1 AGB95406.1 AGB95408.1 CM000162 EDX01757.2 KRK06331.1 KRK06332.1 AJWK01023387 AJWK01023388 AJWK01023389 AJWK01023390 AJWK01023391 AJWK01023392 CH954180 EDV47030.2 AXCN02000407 AY895165 AAW81958.1 GFDL01004149 JAV30896.1 AY902310 AAW82472.1 CH477226 EAT47132.1 AY955097 AAX55681.1 DQ010403 AAY25027.1 JXUM01065716 JXUM01065717 JXUM01065718 JXUM01065719 KQ562361 KXJ76064.1 CH902622 EDV34287.2 ATLV01019151 KE525263 KFB43800.1 GFDL01004151 JAV30894.1 CH480871 EDW53821.1 GGFM01000969 MBW21720.1 GGFJ01001893 MBW51034.1 GFDL01004150 JAV30895.1 CVRI01000042 CRK95719.1 KY660529 AUO38590.1 AAAB01008960 EAA11757.4 OUUW01000003 SPP78695.1 KA645746 AFP60375.1 JRES01001701 KNC20843.1 CH916376 EDV95361.1 GGFK01000909 MBW34230.1 GGFK01000914 MBW34235.1 DQ303468 ABC18327.1 CH963925 EDW78400.2 CH940653 EDW62655.2 KZ149961 PZC76337.1 GGFJ01000980 MBW50121.1 CP012528 ALC48983.1 CH933811 EDW06509.2 GGFL01005920 MBW70098.1 JXJN01000025 CCAG010022741 JX291118 AFQ01087.1 ADMH02001054 ETN64300.1 GGFL01005897 MBW70075.1 APCN01000088 CCAG010020089 JX291119 AFQ01088.1 AGBW02008997 OWR51919.1 GBXI01002085 JAD12207.1 JXJN01012462 AB359912 BAJ05086.1 EU249372 ABY76059.1 GDHF01016734 JAI35580.1 MG763072 AYN61558.1 GAKP01008165 JAC50787.1 JXUM01005967 JXUM01005968 JXUM01005969 JXUM01005970 GBXI01001867 JAD12425.1 GAMC01011138 GAMC01011135 JAB95420.1
ACJ04052.1 MH050927 AZL94115.1 KJ184572 KJ825895 AHX26585.1 AJW29006.1 GDQN01002916 JAT88138.1 ODYU01008646 SOQ52409.1 JTDY01001246 KOB74415.1 KT361877 ANZ54967.1 KX821731 ATB19377.1 AGBW02008921 OWR52183.1 KQ460397 KPJ15233.1 GANO01003978 JAB55893.1 GFDF01008622 JAV05462.1 UFQS01000067 UFQT01000067 SSW98950.1 SSX19332.1 CM000366 EDX17962.1 SSW98949.1 SSX19331.1 GFDF01008259 JAV05825.1 GFDF01008260 JAV05824.1 GFDF01008261 JAV05823.1 ODYU01005061 SOQ45568.1 AXCM01004842 CH379063 EAL32363.3 AE014298 BT132669 AAF48439.2 ADV37675.1 AEQ28953.1 AGB95406.1 AGB95408.1 CM000162 EDX01757.2 KRK06331.1 KRK06332.1 AJWK01023387 AJWK01023388 AJWK01023389 AJWK01023390 AJWK01023391 AJWK01023392 CH954180 EDV47030.2 AXCN02000407 AY895165 AAW81958.1 GFDL01004149 JAV30896.1 AY902310 AAW82472.1 CH477226 EAT47132.1 AY955097 AAX55681.1 DQ010403 AAY25027.1 JXUM01065716 JXUM01065717 JXUM01065718 JXUM01065719 KQ562361 KXJ76064.1 CH902622 EDV34287.2 ATLV01019151 KE525263 KFB43800.1 GFDL01004151 JAV30894.1 CH480871 EDW53821.1 GGFM01000969 MBW21720.1 GGFJ01001893 MBW51034.1 GFDL01004150 JAV30895.1 CVRI01000042 CRK95719.1 KY660529 AUO38590.1 AAAB01008960 EAA11757.4 OUUW01000003 SPP78695.1 KA645746 AFP60375.1 JRES01001701 KNC20843.1 CH916376 EDV95361.1 GGFK01000909 MBW34230.1 GGFK01000914 MBW34235.1 DQ303468 ABC18327.1 CH963925 EDW78400.2 CH940653 EDW62655.2 KZ149961 PZC76337.1 GGFJ01000980 MBW50121.1 CP012528 ALC48983.1 CH933811 EDW06509.2 GGFL01005920 MBW70098.1 JXJN01000025 CCAG010022741 JX291118 AFQ01087.1 ADMH02001054 ETN64300.1 GGFL01005897 MBW70075.1 APCN01000088 CCAG010020089 JX291119 AFQ01088.1 AGBW02008997 OWR51919.1 GBXI01002085 JAD12207.1 JXJN01012462 AB359912 BAJ05086.1 EU249372 ABY76059.1 GDHF01016734 JAI35580.1 MG763072 AYN61558.1 GAKP01008165 JAC50787.1 JXUM01005967 JXUM01005968 JXUM01005969 JXUM01005970 GBXI01001867 JAD12425.1 GAMC01011138 GAMC01011135 JAB95420.1
Proteomes
UP000005204
UP000037510
UP000007151
UP000053240
UP000000304
UP000075900
+ More
UP000075883 UP000075882 UP000001819 UP000000803 UP000075885 UP000002282 UP000076408 UP000092461 UP000008711 UP000075884 UP000075920 UP000075886 UP000075880 UP000069272 UP000008820 UP000069940 UP000249989 UP000007801 UP000030765 UP000001292 UP000183832 UP000007062 UP000268350 UP000037069 UP000001070 UP000007798 UP000008792 UP000192221 UP000095301 UP000092553 UP000095300 UP000009192 UP000091820 UP000092460 UP000092445 UP000092444 UP000078200 UP000000673 UP000092443 UP000075840 UP000076407
UP000075883 UP000075882 UP000001819 UP000000803 UP000075885 UP000002282 UP000076408 UP000092461 UP000008711 UP000075884 UP000075920 UP000075886 UP000075880 UP000069272 UP000008820 UP000069940 UP000249989 UP000007801 UP000030765 UP000001292 UP000183832 UP000007062 UP000268350 UP000037069 UP000001070 UP000007798 UP000008792 UP000192221 UP000095301 UP000092553 UP000095300 UP000009192 UP000091820 UP000092460 UP000092445 UP000092444 UP000078200 UP000000673 UP000092443 UP000075840 UP000076407
Interpro
Gene 3D
ProteinModelPortal
B0LL81
H9J7C4
B6VB33
A0A3Q8UB92
A0A023PPA4
A0A1E1WMG6
+ More
A0A2H1WH64 A0A0L7LG82 A0A1B2INL9 A0A290DSG2 A0A212FEP1 A0A194RBM2 U5ER17 A0A1L8DG79 A0A336KCZ4 B4R561 A0A336LQY1 A0A1L8DHG9 A0A1L8DHK9 A0A1L8DHA0 A0A182RPF2 A0A2H1VXM9 A0A182MNT8 A0A182KPF2 Q29JD8 Q9VXW7 A0A182PJJ1 B4PWL1 A0A182YCW7 A0A1B0CQB9 B3NTM6 A0A182NKC9 A0A182WKJ3 A0A182QYH5 Q5EFD7 A0A182ISC3 A0A1Q3FTU3 A0A182FCQ9 Q5EE11 Q17KB5 Q58FS0 Q4ZH01 A0A182GI95 B3MR82 A0A084W0Q6 A0A1Q3FTH3 B4ILF8 A0A2M3YZM3 A0A2M4BDB1 A0A1Q3FTK2 A0A1J1I5W3 A0A1S4EZU7 A0A2K9P5C3 Q7Q663 A0A3B0J9S9 T1PAE6 A0A0L0BLM0 B4JX85 A0A2M4A0I1 A0A2M4A0G3 Q2PT35 B4N211 B4M788 A0A2W1BSZ9 A0A1W4VFK7 A0A2M4BAV8 A0A1I8NGB4 A0A0M3QZ86 A0A1I8PHG3 B4L5Q0 A0A2M4CZ07 A0A1A9WWZ6 A0A1A9WAP8 A0A1B0ALM4 A0A1B0ABZ5 R4IJP6 A0A1A9ULG0 W5JN31 A0A1A9X7T0 A0A2M4CXT4 A0A182I1Q7 R4IJL6 A0A212FDU0 A0A1A9ZQE4 A0A0A1XMM1 A0A1B0BDG6 A0A1A9YJ64 D4Q9H9 B0LL82 A0A1A9UE24 A0A182X776 A0A0K8VAK9 A0A3G2KQA7 A0A034W909 A0A182HFD7 A0A0A1XNH9 W8B2P5
A0A2H1WH64 A0A0L7LG82 A0A1B2INL9 A0A290DSG2 A0A212FEP1 A0A194RBM2 U5ER17 A0A1L8DG79 A0A336KCZ4 B4R561 A0A336LQY1 A0A1L8DHG9 A0A1L8DHK9 A0A1L8DHA0 A0A182RPF2 A0A2H1VXM9 A0A182MNT8 A0A182KPF2 Q29JD8 Q9VXW7 A0A182PJJ1 B4PWL1 A0A182YCW7 A0A1B0CQB9 B3NTM6 A0A182NKC9 A0A182WKJ3 A0A182QYH5 Q5EFD7 A0A182ISC3 A0A1Q3FTU3 A0A182FCQ9 Q5EE11 Q17KB5 Q58FS0 Q4ZH01 A0A182GI95 B3MR82 A0A084W0Q6 A0A1Q3FTH3 B4ILF8 A0A2M3YZM3 A0A2M4BDB1 A0A1Q3FTK2 A0A1J1I5W3 A0A1S4EZU7 A0A2K9P5C3 Q7Q663 A0A3B0J9S9 T1PAE6 A0A0L0BLM0 B4JX85 A0A2M4A0I1 A0A2M4A0G3 Q2PT35 B4N211 B4M788 A0A2W1BSZ9 A0A1W4VFK7 A0A2M4BAV8 A0A1I8NGB4 A0A0M3QZ86 A0A1I8PHG3 B4L5Q0 A0A2M4CZ07 A0A1A9WWZ6 A0A1A9WAP8 A0A1B0ALM4 A0A1B0ABZ5 R4IJP6 A0A1A9ULG0 W5JN31 A0A1A9X7T0 A0A2M4CXT4 A0A182I1Q7 R4IJL6 A0A212FDU0 A0A1A9ZQE4 A0A0A1XMM1 A0A1B0BDG6 A0A1A9YJ64 D4Q9H9 B0LL82 A0A1A9UE24 A0A182X776 A0A0K8VAK9 A0A3G2KQA7 A0A034W909 A0A182HFD7 A0A0A1XNH9 W8B2P5
PDB
4F3L
E-value=1.77958e-14,
Score=194
Ontologies
GO
Topology
Subcellular location
Nucleus
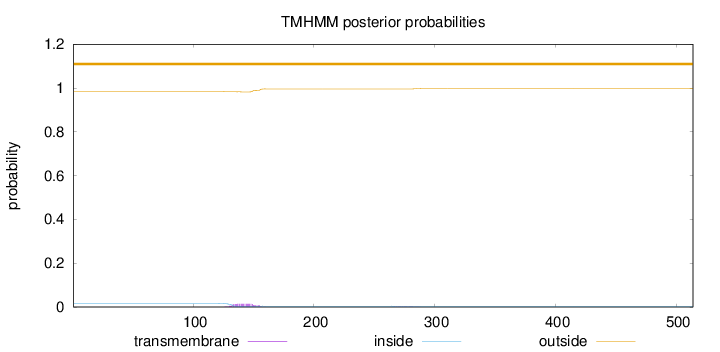
Length:
514
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.34513
Exp number, first 60 AAs:
0.00057
Total prob of N-in:
0.01658
outside
1 - 514
Population Genetic Test Statistics
Pi
335.746745
Theta
235.947065
Tajima's D
1.284565
CLR
0
CSRT
0.736463176841158
Interpretation
Possibly Positive selection
