Gene
KWMTBOMO04365
Pre Gene Modal
BGIBMGA005253
Annotation
PREDICTED:_POU_domain?_class_6?_transcription_factor_2_isoform_X2_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 4.836
Sequence
CDS
ATGAACTTCAGCAATACCAGGGGCAGAGCCAAGCCGCTGCCTACCGCTTGGCAGGCGGAATCGGATCGGGAGAGTGGCGCGAGTTCGCCGGAGGGGGGCACGACGCGGGCGCAATTAGCTGAGCCGCGTACCCCCTCGCCAAGAACCAGGACACCGCACCACCAGCACATGAATGGCTCCATGGCGTCGATGTTCCAAAACCTGCAGAACCTCGCGAGCATGCAACAGAACATGCCTATGTCGCAACAACTCCAGCAGCAGATGTCCCAGCAGATGTCGCAGCTGGCGGCAAATCTCCAGGGTCTGGCGTCCATGCCGTCGAATCCGGTTATCAACTCCCCTTTAAACTTGAGCGTCAGCGCCCCGGGAATGGGCTCCCCGAGCCCCGTGAGCAACAACAATATGCTGCCACCAGCGATGCCGTCTCCGATGCCGCAGCTAATACTCGCGTCAGGGCAACTGGTCCAAGGAATCCAGGGTGCTCAACTACTCATACCTACTTCGCAAGGAATCGCAACCCAAACAATCCTGACGATACCAGTCAACCACGTCAACTCTAACGATCAAATGGTGAACCTAGCTCTAAACAACGGCCAAGTCGTCTCCACGTCCCTAGCCAATCTCCAAGCGATGGCCCAACCCCATCAGCTACTCAACTCGAATCCCCAACAATCATCAAACATTCGACCCAACATGCTGAATCCAAGTTTATCGAATGCCCTACTCAACCCGGGCTTACCGAACTTCTTGTCTAACGGAGCGGCCAATGCCCAAGAATTGCTTCAAGCGTTACAACAACAGCAGCCTCAAGGGAACCACAGTTTGCTGCAGACGGTACAGCAGAACAATATGGCGCAGCAGATGCAAGGCAGAAGATCCTCTTCTCCAAGGCCTGAGAGGCATTACAAAGAGAGGGAGAGCTACGAACGGTATTCTAGTAGTGTTAGAGAGAGAAATGAACGTGAGGGAGCAGCAGCCATGAACAGTATTAATAGGCTTGCTGCGTCGAATGGCGAGATCACGATCACGACGTCCCACAACGCGAGCGGCGCGACTAGCAGCGCCGGCAGCGGAACCGGGCCGATTCCGTCGCCGCACGCACCCGTCAAACTATCGCCGGGCTCGGTGAAGTCGCCATCGCACGACGACGCCGATCTGTTGGCAGACTCACCAAACCAGCCAACGATCAGCCAGTCTTCCAGCAACGTAGTCGATGGGATCAACTTAGAAGACATCAAGGAATTCGCGAAGGCCTTCAAACTGCGACGACTTGGTCTGGGACTTACGCAGACCCAAGTAGGCCAAGCCCTGTCGGTCACCGAAGGACCAGCATATAGTCAAAGTGCGATCTGCAGTGCCCTGGCTTCGCAGATGCTAGCAGCTCAACTGTCTTCACAACAACAAAACATATTTGAGAAATTGGATATAACACCAAAAAGTGCGCAGAAAATCAAACCGGTGCTCGAACGTTGGATGAAGGAAGCCGAGGAAAGGTACGCATCCGGTCAAAATCATCTGACGGACTTCATTGGAATGGAGCCGAGCAAAAAGCGCAAACGGCGAACATCGTTCACACCTCAGGCCCTAGAACTGCTCAACGCGCACTTCGAAAGGAACACCCATCCATCTGGCACTGAAATAACCGGACTTGCCCATCAGTTAGGCTACGAACGAGAGGTCATCAGAATATGGTTTTGCAACAAGCGACAAGCTCTCAAGAACACTGTCCGGATGATGTCCAAAGGAATGGTCTAA
Protein
MNFSNTRGRAKPLPTAWQAESDRESGASSPEGGTTRAQLAEPRTPSPRTRTPHHQHMNGSMASMFQNLQNLASMQQNMPMSQQLQQQMSQQMSQLAANLQGLASMPSNPVINSPLNLSVSAPGMGSPSPVSNNNMLPPAMPSPMPQLILASGQLVQGIQGAQLLIPTSQGIATQTILTIPVNHVNSNDQMVNLALNNGQVVSTSLANLQAMAQPHQLLNSNPQQSSNIRPNMLNPSLSNALLNPGLPNFLSNGAANAQELLQALQQQQPQGNHSLLQTVQQNNMAQQMQGRRSSSPRPERHYKERESYERYSSSVRERNEREGAAAMNSINRLAASNGEITITTSHNASGATSSAGSGTGPIPSPHAPVKLSPGSVKSPSHDDADLLADSPNQPTISQSSSNVVDGINLEDIKEFAKAFKLRRLGLGLTQTQVGQALSVTEGPAYSQSAICSALASQMLAAQLSSQQQNIFEKLDITPKSAQKIKPVLERWMKEAEERYASGQNHLTDFIGMEPSKKRKRRTSFTPQALELLNAHFERNTHPSGTEITGLAHQLGYEREVIRIWFCNKRQALKNTVRMMSKGMV
Summary
Similarity
Belongs to the POU transcription factor family.
Uniprot
Pubmed
EMBL
Proteomes
Interpro
ProteinModelPortal
PDB
3D1N
E-value=1.70506e-51,
Score=514
Ontologies
GO
Topology
Subcellular location
Nucleus
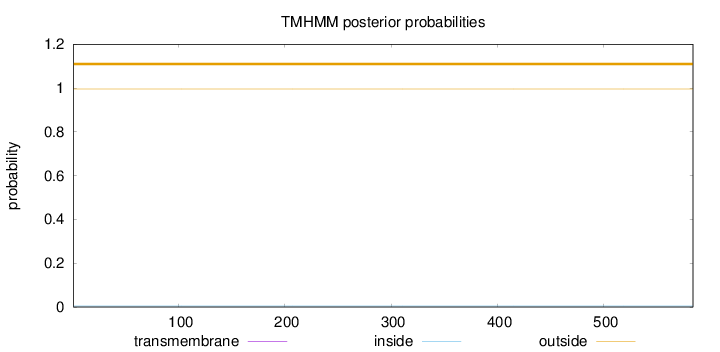
Length:
584
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00566999999999999
Exp number, first 60 AAs:
7e-05
Total prob of N-in:
0.00462
outside
1 - 584
Population Genetic Test Statistics
Pi
209.877792
Theta
165.169892
Tajima's D
0.84253
CLR
0.330333
CSRT
0.611169441527924
Interpretation
Uncertain
