Gene
KWMTBOMO04041
Pre Gene Modal
BGIBMGA010283
Annotation
PREDICTED:_septin_and_tuftelin_interacting_protein_isoform_X1_[Bombyx_mori]
Full name
Septin-interacting protein 1
Transcription factor
Location in the cell
Nuclear Reliability : 3.867
Sequence
CDS
ATGTCAGACGATGAGGTTATAAAGTTTGAGATCACAGACTACGATTTGGATAATGAATTCAACCCCGGCCGTGGAAGAAAAGCTAACAAGGAACAACAAATATACGGTGTATGGGCCAAAGACAGTGATGATGATGAAAATGAAGATAATGTTAGACAGAGAATACGTAAACCTAAAGATTTTTCTGCACCAATTGGTTTCGTGGCGGGTGGAGTTCAACAGGCTGGCAAAAAAAAAGACAAGACTAAAGAAATAAAACCAGCTGAAGCGTCTACTTCTGCTCCTAAGTACTCAGACAGTTCTGATGAGGAACCAGAGGTTCCAGTAGCGAGTGACACGGCGGGAATCAGGAGACAGGGACAGGGAATGCGTCCACCATCATTAGGTGGAAATGTGGGAAATTGGGAAAAACATACTAAAGGAATCGGAGCAAAATTGTTGTTGCAGATGGGATACCAACCGGGTAAAGGTTTGGGTAAAGATTTGCAAGGTATCTCAGCGCCGGTCGAGGCTACGCTGCGCAAAGGCAGAGGTGCGATCGGAGCTTACGGACCAGAAAAGGCAACGATAAAAGCCAAAAAAGAAGAGGAACGTCGCATCAAAGATAAAGAAGATCAGGATAAAGATAAAACCGAAAAGAATTACAACTGGAAGAAATCGCACAAGGGTCGTTATTTTTACAGAGATGCCGCTGACGTCATTCAGGAGGGTAAACCGACAACGTATACCATAGCAAGTAACGAGTTGTCCCGCGTGCCCGTGATCGACATGACCGGCAAGGAGAAGCGCGTGCTGAGCGGCTACCACGCGCTGCGGGCGCAGGCGCCGCGCTACGAGCACGAGCCCAGGAGGAAGTGCCAGAACTTCTCCGCGCCGCAGCTGGTCCACAACCTCGAGGTCATGGTCGAGTGCTGCGAACAGGTCCAAGCCCTCAGGTTTATACATCTTTATCGCTTAGACAAGTGGTGGAATTCACAGCCGACCTGGTGTTAA
Protein
MSDDEVIKFEITDYDLDNEFNPGRGRKANKEQQIYGVWAKDSDDDENEDNVRQRIRKPKDFSAPIGFVAGGVQQAGKKKDKTKEIKPAEASTSAPKYSDSSDEEPEVPVASDTAGIRRQGQGMRPPSLGGNVGNWEKHTKGIGAKLLLQMGYQPGKGLGKDLQGISAPVEATLRKGRGAIGAYGPEKATIKAKKEEERRIKDKEDQDKDKTEKNYNWKKSHKGRYFYRDAADVIQEGKPTTYTIASNELSRVPVIDMTGKEKRVLSGYHALRAQAPRYEHEPRRKCQNFSAPQLVHNLEVMVECCEQVQALRFIHLYRLDKWWNSQPTWC
Summary
Description
May be involved in pre-mRNA splicing.
Similarity
Belongs to the TFP11/STIP family.
Uniprot
A1XDB3
H9JL82
A0A212FCY7
A0A194RKK6
A0A194Q5P5
A0A2A4K201
+ More
A0A2H1WJM9 A0A2W1BUW5 A0A1E1VZP1 S4PDH3 A0A139WJU9 A0A1W4WLB1 A1XDA7 A0A1Y1LE60 A0A1Y1LAH6 N6TNF2 A0A1Y1LGA8 A0A1B0GIT1 A0A182R3Y2 A0A182NAK3 A0A1Y1LAH9 A0A182Q9K9 Q16YS0 A0A1B0D5Y9 A0A1Y1LAH8 A0A182KWD4 A0A182TUL0 A0A182F3B0 A1XDB4 A0A182VTE4 A0A1S4G738 A0A182GJF2 C6ETQ3 A0A182LTI5 Q16YS1 A0A3Q0IP11 A0A182IBE0 W5JQZ1 A0A182X6B7 A1XDB5 A0A182VK21 A0A182PIZ5 A0A084VME3 C6ETS6 A0A146KZW4 A0A182YR69 B0W0H4 A0A182SHH9 A0A182J5M6 T1IB51 A0A0A9X2E3 C6ETS8 U4U4Z9 C6ETQ5 C6ETS9 A0A034WBN6 A0A1B6MLQ3 C6ETQ4 W8BBQ4 C6ETS5 C6ETR8 A0A1Q3F2N3 A0A182JXN2 A0A0K8VF45 A0A0C9QGZ0 A0A1Y1LAG5 A0A1A9USN8 A0A0M9AA34 A0A023F277 A0A151J015 A0A1B6IUK7 A0A0K8SVR8 A0A0J7KL79 C6ETR1 A0A154PBQ0 E2BJK8 B4KGZ0 C6ETS1 A0A1Y1LE80 F4WE40 A0A0L7RJW0 A0A158NUU0 A0A1B0A3T9 A0A195BBD2 C6ETS0 A0A151X453 A0A0L0BWZ6 A0A151K073 A0A1B0G097 A0A2S2Q3N6 E0VXQ2 A0A2S2PIW7 A0A310SNI8 A1XDA8 A0A2A3EJI9 A0A1A9X057 A0A1J1I0P0 A0A1B6GH69 J9JQN9
A0A2H1WJM9 A0A2W1BUW5 A0A1E1VZP1 S4PDH3 A0A139WJU9 A0A1W4WLB1 A1XDA7 A0A1Y1LE60 A0A1Y1LAH6 N6TNF2 A0A1Y1LGA8 A0A1B0GIT1 A0A182R3Y2 A0A182NAK3 A0A1Y1LAH9 A0A182Q9K9 Q16YS0 A0A1B0D5Y9 A0A1Y1LAH8 A0A182KWD4 A0A182TUL0 A0A182F3B0 A1XDB4 A0A182VTE4 A0A1S4G738 A0A182GJF2 C6ETQ3 A0A182LTI5 Q16YS1 A0A3Q0IP11 A0A182IBE0 W5JQZ1 A0A182X6B7 A1XDB5 A0A182VK21 A0A182PIZ5 A0A084VME3 C6ETS6 A0A146KZW4 A0A182YR69 B0W0H4 A0A182SHH9 A0A182J5M6 T1IB51 A0A0A9X2E3 C6ETS8 U4U4Z9 C6ETQ5 C6ETS9 A0A034WBN6 A0A1B6MLQ3 C6ETQ4 W8BBQ4 C6ETS5 C6ETR8 A0A1Q3F2N3 A0A182JXN2 A0A0K8VF45 A0A0C9QGZ0 A0A1Y1LAG5 A0A1A9USN8 A0A0M9AA34 A0A023F277 A0A151J015 A0A1B6IUK7 A0A0K8SVR8 A0A0J7KL79 C6ETR1 A0A154PBQ0 E2BJK8 B4KGZ0 C6ETS1 A0A1Y1LE80 F4WE40 A0A0L7RJW0 A0A158NUU0 A0A1B0A3T9 A0A195BBD2 C6ETS0 A0A151X453 A0A0L0BWZ6 A0A151K073 A0A1B0G097 A0A2S2Q3N6 E0VXQ2 A0A2S2PIW7 A0A310SNI8 A1XDA8 A0A2A3EJI9 A0A1A9X057 A0A1J1I0P0 A0A1B6GH69 J9JQN9
Pubmed
EMBL
DQ342046
ABC69938.1
BABH01011772
BABH01011773
AGBW02009118
OWR51615.1
+ More
KQ460075 KPJ17954.1 KQ459582 KPI98720.1 NWSH01000282 PCG77793.1 ODYU01009050 SOQ53176.1 KZ149930 PZC77424.1 GDQN01010856 JAT80198.1 GAIX01003676 JAA88884.1 KQ971338 KYB28202.1 DQ342040 ABC69932.1 KYB28201.1 GEZM01061225 GEZM01061218 GEZM01061217 JAV70630.1 GEZM01061224 GEZM01061222 GEZM01061219 GEZM01061214 JAV70634.1 APGK01057945 APGK01057946 KB741283 ENN70775.1 GEZM01061221 JAV70636.1 AJWK01015376 GEZM01061216 JAV70649.1 AXCN02002191 CH477510 EAT39779.1 AJVK01003470 GEZM01061223 JAV70633.1 DQ342047 ABC69939.1 JXUM01012806 KQ560362 KXJ82820.1 EU707401 ACJ45811.1 AXCM01004755 EAT39778.1 APCN01000672 ADMH02000364 ETN66817.1 AAAB01008944 DQ342048 HACL01000127 ABC69940.1 CFW94421.1 ATLV01014586 KE524975 KFB39137.1 EU707425 ACJ45836.1 GDHC01018087 JAQ00542.1 DS231817 EDS39713.1 ACPB03007249 GBHO01029490 JAG14114.1 EU707427 ACJ45837.1 KB631738 ERL85681.1 EU707403 ACJ45813.1 EU707428 ACJ45832.1 GAKP01005951 JAC53001.1 GEBQ01003089 JAT36888.1 EU707402 ACJ45812.1 GAMC01007895 JAB98660.1 EU707424 ACJ45835.1 EU707417 ACJ45834.1 GFDL01013258 JAV21787.1 GDHF01014831 JAI37483.1 GBYB01002744 JAG72511.1 GEZM01061220 JAV70639.1 KQ435704 KOX80307.1 GBBI01003521 JAC15191.1 KQ980670 KYN14624.1 GECU01017098 JAS90608.1 GBRD01008428 JAG57393.1 LBMM01006021 KMQ91009.1 EU707409 ACJ45819.1 KQ434869 KZC09329.1 GL448571 EFN84165.1 CH933807 EDW12201.1 EU707420 ACJ45827.1 GEZM01061215 JAV70650.1 GL888102 EGI67478.1 KQ414579 KOC71124.1 ADTU01026715 KQ976532 KYM81525.1 EU707419 ACJ45826.1 KQ982557 KYQ55054.1 JRES01001211 KNC24562.1 KQ981296 KYN43118.1 CCAG010018487 GGMS01003028 MBY72231.1 DS235833 EEB18158.1 GGMR01016783 MBY29402.1 KQ759784 OAD62855.1 DQ342041 ABC69933.1 KZ288238 PBC31346.1 CVRI01000038 CRK93763.1 GECZ01008655 GECZ01008002 JAS61114.1 JAS61767.1 ABLF02036507
KQ460075 KPJ17954.1 KQ459582 KPI98720.1 NWSH01000282 PCG77793.1 ODYU01009050 SOQ53176.1 KZ149930 PZC77424.1 GDQN01010856 JAT80198.1 GAIX01003676 JAA88884.1 KQ971338 KYB28202.1 DQ342040 ABC69932.1 KYB28201.1 GEZM01061225 GEZM01061218 GEZM01061217 JAV70630.1 GEZM01061224 GEZM01061222 GEZM01061219 GEZM01061214 JAV70634.1 APGK01057945 APGK01057946 KB741283 ENN70775.1 GEZM01061221 JAV70636.1 AJWK01015376 GEZM01061216 JAV70649.1 AXCN02002191 CH477510 EAT39779.1 AJVK01003470 GEZM01061223 JAV70633.1 DQ342047 ABC69939.1 JXUM01012806 KQ560362 KXJ82820.1 EU707401 ACJ45811.1 AXCM01004755 EAT39778.1 APCN01000672 ADMH02000364 ETN66817.1 AAAB01008944 DQ342048 HACL01000127 ABC69940.1 CFW94421.1 ATLV01014586 KE524975 KFB39137.1 EU707425 ACJ45836.1 GDHC01018087 JAQ00542.1 DS231817 EDS39713.1 ACPB03007249 GBHO01029490 JAG14114.1 EU707427 ACJ45837.1 KB631738 ERL85681.1 EU707403 ACJ45813.1 EU707428 ACJ45832.1 GAKP01005951 JAC53001.1 GEBQ01003089 JAT36888.1 EU707402 ACJ45812.1 GAMC01007895 JAB98660.1 EU707424 ACJ45835.1 EU707417 ACJ45834.1 GFDL01013258 JAV21787.1 GDHF01014831 JAI37483.1 GBYB01002744 JAG72511.1 GEZM01061220 JAV70639.1 KQ435704 KOX80307.1 GBBI01003521 JAC15191.1 KQ980670 KYN14624.1 GECU01017098 JAS90608.1 GBRD01008428 JAG57393.1 LBMM01006021 KMQ91009.1 EU707409 ACJ45819.1 KQ434869 KZC09329.1 GL448571 EFN84165.1 CH933807 EDW12201.1 EU707420 ACJ45827.1 GEZM01061215 JAV70650.1 GL888102 EGI67478.1 KQ414579 KOC71124.1 ADTU01026715 KQ976532 KYM81525.1 EU707419 ACJ45826.1 KQ982557 KYQ55054.1 JRES01001211 KNC24562.1 KQ981296 KYN43118.1 CCAG010018487 GGMS01003028 MBY72231.1 DS235833 EEB18158.1 GGMR01016783 MBY29402.1 KQ759784 OAD62855.1 DQ342041 ABC69933.1 KZ288238 PBC31346.1 CVRI01000038 CRK93763.1 GECZ01008655 GECZ01008002 JAS61114.1 JAS61767.1 ABLF02036507
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000218220
UP000007266
+ More
UP000192223 UP000019118 UP000092461 UP000075900 UP000075884 UP000075886 UP000008820 UP000092462 UP000075882 UP000075902 UP000069272 UP000075920 UP000069940 UP000249989 UP000075883 UP000079169 UP000075840 UP000000673 UP000076407 UP000075903 UP000075885 UP000030765 UP000076408 UP000002320 UP000075901 UP000075880 UP000015103 UP000030742 UP000075881 UP000078200 UP000053105 UP000078492 UP000036403 UP000076502 UP000008237 UP000009192 UP000007755 UP000053825 UP000005205 UP000092445 UP000078540 UP000075809 UP000037069 UP000078541 UP000092444 UP000009046 UP000005203 UP000242457 UP000091820 UP000183832 UP000007819
UP000192223 UP000019118 UP000092461 UP000075900 UP000075884 UP000075886 UP000008820 UP000092462 UP000075882 UP000075902 UP000069272 UP000075920 UP000069940 UP000249989 UP000075883 UP000079169 UP000075840 UP000000673 UP000076407 UP000075903 UP000075885 UP000030765 UP000076408 UP000002320 UP000075901 UP000075880 UP000015103 UP000030742 UP000075881 UP000078200 UP000053105 UP000078492 UP000036403 UP000076502 UP000008237 UP000009192 UP000007755 UP000053825 UP000005205 UP000092445 UP000078540 UP000075809 UP000037069 UP000078541 UP000092444 UP000009046 UP000005203 UP000242457 UP000091820 UP000183832 UP000007819
ProteinModelPortal
A1XDB3
H9JL82
A0A212FCY7
A0A194RKK6
A0A194Q5P5
A0A2A4K201
+ More
A0A2H1WJM9 A0A2W1BUW5 A0A1E1VZP1 S4PDH3 A0A139WJU9 A0A1W4WLB1 A1XDA7 A0A1Y1LE60 A0A1Y1LAH6 N6TNF2 A0A1Y1LGA8 A0A1B0GIT1 A0A182R3Y2 A0A182NAK3 A0A1Y1LAH9 A0A182Q9K9 Q16YS0 A0A1B0D5Y9 A0A1Y1LAH8 A0A182KWD4 A0A182TUL0 A0A182F3B0 A1XDB4 A0A182VTE4 A0A1S4G738 A0A182GJF2 C6ETQ3 A0A182LTI5 Q16YS1 A0A3Q0IP11 A0A182IBE0 W5JQZ1 A0A182X6B7 A1XDB5 A0A182VK21 A0A182PIZ5 A0A084VME3 C6ETS6 A0A146KZW4 A0A182YR69 B0W0H4 A0A182SHH9 A0A182J5M6 T1IB51 A0A0A9X2E3 C6ETS8 U4U4Z9 C6ETQ5 C6ETS9 A0A034WBN6 A0A1B6MLQ3 C6ETQ4 W8BBQ4 C6ETS5 C6ETR8 A0A1Q3F2N3 A0A182JXN2 A0A0K8VF45 A0A0C9QGZ0 A0A1Y1LAG5 A0A1A9USN8 A0A0M9AA34 A0A023F277 A0A151J015 A0A1B6IUK7 A0A0K8SVR8 A0A0J7KL79 C6ETR1 A0A154PBQ0 E2BJK8 B4KGZ0 C6ETS1 A0A1Y1LE80 F4WE40 A0A0L7RJW0 A0A158NUU0 A0A1B0A3T9 A0A195BBD2 C6ETS0 A0A151X453 A0A0L0BWZ6 A0A151K073 A0A1B0G097 A0A2S2Q3N6 E0VXQ2 A0A2S2PIW7 A0A310SNI8 A1XDA8 A0A2A3EJI9 A0A1A9X057 A0A1J1I0P0 A0A1B6GH69 J9JQN9
A0A2H1WJM9 A0A2W1BUW5 A0A1E1VZP1 S4PDH3 A0A139WJU9 A0A1W4WLB1 A1XDA7 A0A1Y1LE60 A0A1Y1LAH6 N6TNF2 A0A1Y1LGA8 A0A1B0GIT1 A0A182R3Y2 A0A182NAK3 A0A1Y1LAH9 A0A182Q9K9 Q16YS0 A0A1B0D5Y9 A0A1Y1LAH8 A0A182KWD4 A0A182TUL0 A0A182F3B0 A1XDB4 A0A182VTE4 A0A1S4G738 A0A182GJF2 C6ETQ3 A0A182LTI5 Q16YS1 A0A3Q0IP11 A0A182IBE0 W5JQZ1 A0A182X6B7 A1XDB5 A0A182VK21 A0A182PIZ5 A0A084VME3 C6ETS6 A0A146KZW4 A0A182YR69 B0W0H4 A0A182SHH9 A0A182J5M6 T1IB51 A0A0A9X2E3 C6ETS8 U4U4Z9 C6ETQ5 C6ETS9 A0A034WBN6 A0A1B6MLQ3 C6ETQ4 W8BBQ4 C6ETS5 C6ETR8 A0A1Q3F2N3 A0A182JXN2 A0A0K8VF45 A0A0C9QGZ0 A0A1Y1LAG5 A0A1A9USN8 A0A0M9AA34 A0A023F277 A0A151J015 A0A1B6IUK7 A0A0K8SVR8 A0A0J7KL79 C6ETR1 A0A154PBQ0 E2BJK8 B4KGZ0 C6ETS1 A0A1Y1LE80 F4WE40 A0A0L7RJW0 A0A158NUU0 A0A1B0A3T9 A0A195BBD2 C6ETS0 A0A151X453 A0A0L0BWZ6 A0A151K073 A0A1B0G097 A0A2S2Q3N6 E0VXQ2 A0A2S2PIW7 A0A310SNI8 A1XDA8 A0A2A3EJI9 A0A1A9X057 A0A1J1I0P0 A0A1B6GH69 J9JQN9
PDB
5Y88
E-value=1.4606e-05,
Score=115
Ontologies
GO
Topology
Subcellular location
Nucleus
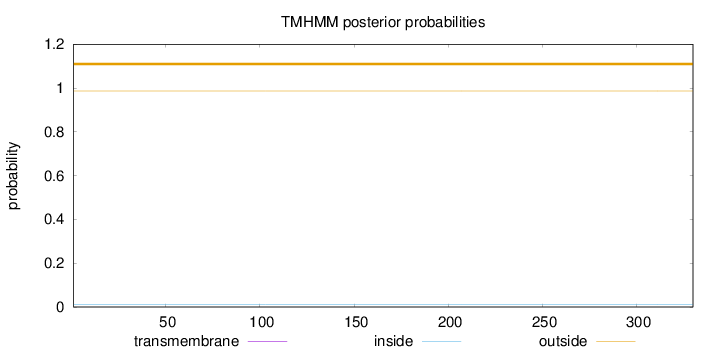
Length:
330
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00176
Exp number, first 60 AAs:
3e-05
Total prob of N-in:
0.01305
outside
1 - 330
Population Genetic Test Statistics
Pi
303.209027
Theta
165.512419
Tajima's D
3.257752
CLR
0.116296
CSRT
0.988650567471626
Interpretation
Uncertain
