Gene
KWMTBOMO04021
Pre Gene Modal
BGIBMGA010110
Annotation
PREDICTED:_mothers_against_decapentaplegic_homolog_4_isoform_X1_[Bombyx_mori]
Full name
Mothers against decapentaplegic homolog
Alternative Name
SMAD family member
Transcription factor
Location in the cell
Nuclear Reliability : 4.247
Sequence
CDS
ATGAATACGACAGCACCAACATCGGCAGATGCATGCCTCAGCATTGTGCATTCACTGATGTGCCACCGTCAAGGTGGTGAGAGCGAAGGATTTTCTAAACGAGCCATTGAATCTCTGGTCAAGAAGTTGAAAGAAAAACGAGATGAGCTTGACTCTTTGATCACAGCTATAACTACAAATGGTGCACATCCTAGTAAATGTGTTACTATACAGAGGACTTTAGATGGGAGATTACAGGTTGCTGGTCGGAAAGGATTTCCTCATGTAATTTATGCTCGAATATGGAGGTGGCCAGATTTACATAAAAATGAACTGAAACATGTTAAATTTTGTCAATTTGCATTTGATTTAAAGTGTGACTCTGTGTGTGTTAATCCATACCACTATGAAAGAGTTGTTTCTCCTGGTATAGATTTATCCGGCTTGTCCCTTCAATCTGGACCCAGTAGATTAGTAAAGGACGAGTACACGGCTGGTCTAGGTGGTAACGGTATGGACATGGATACTGGTGAGATGGTTACCATACAGCATCATGCGACAAGTCCTAGACACCACCATTCGACAATACAACATCACCATCAACAGTTTCAAACAACTAATATTATTATGAATCAAGGACAGACGCCCGATGGCGTTCCGAGCATGTTTTCACCGACCCACGGGCCCCGGACGCCGATCCGGCCTTCGGGTGGTCCATCCCAGTTACCACCGCCGCCTTCCATGGGACACTCGCCGAGCGCACAAATGATGCAAAATCACACGGGACAAATGCCCACACCGCAGATGGTACCGGGGGCTTCACAACTACGATCGGGAACGCCCCAAATGGGACCCGCTACGCCACAAATGGTCCCCGGAACACCACAGATGGTGCCCGGTAATCCGCAAATGGTACCGAACACCCCGCAAATGGGGCCGGGCACACCGCAGATGGGTCCCAGTACTCCGCAAATGGGTCCGGGCACGCCACAGATGGGACCCGGTACACCTCAGATGGGTCCGGGTACACCTCAGATGGGATCAAATGTACCTCAAATGGCTTCGCCGAGAATGGCTTCGGCCCCCACGCAGATGTCTCCGGGGACCCCCCAGATGCCTTCGATTAGTCAGGGAATGTCCATACCGAGCCCGCAACAAATGGTGATGCAGCAAAGAGCCATAGCCCCTAAGCTGGAGCCACCCGACACCATGGACGCCCGAGCTATGTGGCTGCCAAAAAGAATGAATCATTCGGCGATACCAGTAAGTATGTCGCCAGGCGCGACGACGCCTCTTATCGATGGTGCGAACACGACGTTCTTCGGCACAGAACAAACCACTACCGACACACAAATCACACAGACTCTCCCTACTGGCACAGTTTCACCGATAACCGGTGAGGGTACACAACAGAACGGGTTTGCTGCGACCAGTCCAGCACCGCAACCTAGTCCTCTTCAGCACCAGAGCCAACCACAGCCACCTCAGCAGCAACAACAACAACAGCAGCAGCAGGGTAACAAGTCATTTTCTTTTTGA
Protein
MNTTAPTSADACLSIVHSLMCHRQGGESEGFSKRAIESLVKKLKEKRDELDSLITAITTNGAHPSKCVTIQRTLDGRLQVAGRKGFPHVIYARIWRWPDLHKNELKHVKFCQFAFDLKCDSVCVNPYHYERVVSPGIDLSGLSLQSGPSRLVKDEYTAGLGGNGMDMDTGEMVTIQHHATSPRHHHSTIQHHHQQFQTTNIIMNQGQTPDGVPSMFSPTHGPRTPIRPSGGPSQLPPPPSMGHSPSAQMMQNHTGQMPTPQMVPGASQLRSGTPQMGPATPQMVPGTPQMVPGNPQMVPNTPQMGPGTPQMGPSTPQMGPGTPQMGPGTPQMGPGTPQMGSNVPQMASPRMASAPTQMSPGTPQMPSISQGMSIPSPQQMVMQQRAIAPKLEPPDTMDARAMWLPKRMNHSAIPVSMSPGATTPLIDGANTTFFGTEQTTTDTQITQTLPTGTVSPITGEGTQQNGFAATSPAPQPSPLQHQSQPQPPQQQQQQQQQQGNKSFSF
Summary
Similarity
Belongs to the dwarfin/SMAD family.
EMBL
Proteomes
Interpro
IPR013019
MAD_homology_MH1
+ More
IPR001132 SMAD_dom_Dwarfin-type
IPR017855 SMAD-like_dom_sf
IPR013790 Dwarfin
IPR008984 SMAD_FHA_dom_sf
IPR036578 SMAD_MH1_sf
IPR003619 MAD_homology1_Dwarfin-type
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR036322 WD40_repeat_dom_sf
IPR001132 SMAD_dom_Dwarfin-type
IPR017855 SMAD-like_dom_sf
IPR013790 Dwarfin
IPR008984 SMAD_FHA_dom_sf
IPR036578 SMAD_MH1_sf
IPR003619 MAD_homology1_Dwarfin-type
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR036322 WD40_repeat_dom_sf
Gene 3D
ProteinModelPortal
PDB
3QSV
E-value=3.02305e-65,
Score=632
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
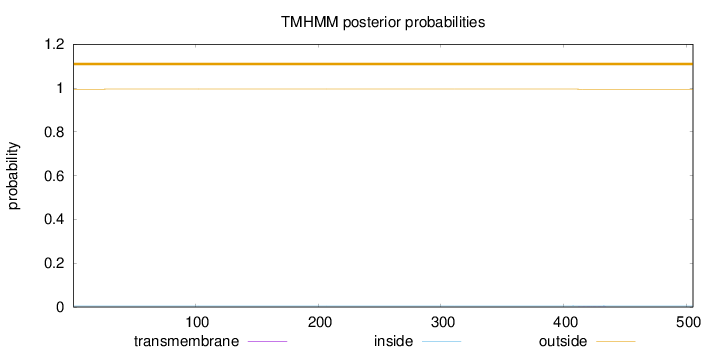
Length:
505
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00478
Exp number, first 60 AAs:
0.0005
Total prob of N-in:
0.00496
outside
1 - 505
Population Genetic Test Statistics
Pi
273.142973
Theta
205.829823
Tajima's D
1.256457
CLR
0
CSRT
0.731713414329283
Interpretation
Uncertain
