Gene
KWMTBOMO03524
Pre Gene Modal
BGIBMGA013750
Annotation
reverse_transcriptase_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.422
Sequence
CDS
ATGGACGCTGTATTTGCGGAATTTCTCCGGCTTCGCTACCCAAAGCTCACTTCCGAGTTTCAGGCCTTCAAGGCCGATCACACCGCGAGCCCTCTCGTGGACTCCACCGAACTCGCTGCTCCTGCGTCGCCTGTACCTGCGTGCAAAGCTTCTGCATCGAGCACCGCAGCCTCCGTCGTGCCTGCCGTTCCCGCGTCGCCTATACTGGCAAGTAAAACTGCTGCGTCGTCCGCCGTGGTCACCGTTCCAGCAGCGCGATCATCCGCGGCCTCCGTCGCGCCCTCCAAAACTCCCGCTCGTAGGTCACCTGCGCCCGCCTCCTCGTCCTCCGACTCTGACTCGGAGATGGAGGTCGACCTCGCCCCCGCCTCATCGACGGATGAATTCACCCTGGTACAAAAGGGTAAGAAGCGTGCCGCGGAGTCTCGAGCTCCCGCGGCCGCTAAAATTAGCAAAGCCGCGAACGCGTCGCGTCCCCGCCCTCAGACTCCCGTTGCGCCTCCAGCCCGTGCCACTCCGTCGCCGCGTCCGGTGGCACAAAAAAAAACCCAGACCCCTCCCCCGGTAATCCTTCAAGAGAAGGCAGCTTGGGATCGAGTTTCCCTGGCCCTTAAGGCCAACAATATCAATTTCACGAATGCCCGTAACCTCGCGAACGGCATTCAAATTAAGGTTCGAACACCCGACGACCATAGGGCCCTCTCTTCTTACCTCCGTAAGGAGCGTATAAGTTTCCACACGTATACGCTCCAGGAGGAGCGCGAACTCAATGCCGTCATACGTGGCATCCCTAAAGAGTTGGATGCCGAGCTCGTCAAGGCCGACCTTCTCGAACAAGGCCTACCGGTTAACTCCGTACACCGCATGCACACAGGCCGCGGAAGGGAGCCATATAACTGATTTTTAAACGCTTACGGTCTCGCAAATCAACGTGATCAGGTTTCTGACTTTTTGCGTGACCATCAAATTGATATCTTTTTAGTGCAGGAGACCCTACTTAAGCCCGCGCGCCGTGACCCTAAAATCGCGAACTATAACATGGTCAGGAACAACAGGCTCTCTGCTCGTGGTGGTACCGTAATTTACTATAGAAGAGCCCTGCATTGCGTCCCGCTCGATCCTCCCGCGCTCGAAGCATCAGTGTGCCGAATCTCACTGACGGGACACGCGCCGATCGTTATCGCGTCCGTTTATCTTCCACCGGATAAGATCGTTCTAAGCAGTGATATCGAGGCGCTGCTCGGCATGGGGAGCTCTGTCATTCTGGCGGGTGACCTAAATTGTAAACATATCAGGTGGAACTCACACACCACAACCCCGAATGGCAGGCGGCTTGACGCATTAGTCGATGATCTCGCCTTCGATATCGTCGCTCCGCTAACCCCGACTCACTACCCGCTAAATATCGCGCATCGCCCGGATATACTCGACATAGCGTTATTAAAAAACGTAACTCTGCGCTTACACTCGATCGAAGTAGTTTCAGAGTTAGATTCAGACCACCGTCCCGTCGTTATGAAGCTCGGTCGCGCTCCCGATTCCGTTCCCGTCACGAGGACTGTGGTGGATTGGCACACGCAGGGCATCAGCCTGGCTGAATCTGATCCACCATCACTCCCGTTTAGCCCGGACTCTACCCCGTCTCCTCAGGATACCGCTGAAGCCATAGACATCTTAACGTCACACATCACCTCGACATTAGATAGGTCATCGAAACAAGTTGTAGCGGAGGACTTCCTTCTCCGCTTCAAATTGTCCGACGATATAGGGAACTCCTTAGAGCTAAAAACGCCTCGATACGCGCCTACGACAGGCCGACTCGCGGCGTTCGATGATGACGAAAAAGCAGAGCTGCTGGCCGATACATTGCAAACCCAGTGCACGCCCAGCACTCAATCCGTGGACCCTGTTCATGTAGAATTAGTAGACAGTGAGGTAGAACGCAGAGCCTCCTTGCCACCCTCGGATGCGTTACCACCCGTCACCCCGATGGAAGTTAAAGACTTGATCAAAGACCTACGTCCTCGCAAGGCTCCCGGTTCCAACGGTATATCCAACCGCGTTATTAAACTTCTACCCGTCCAAGTCATCGTGATGTTGGCATCTATTTTCAATGCCGCTATGACGAACTGTATCTTTCCCGCGGTGTGGAAAGAAGCGGACGTTATCGGCATACATAAACCCGGTAAACCAAAAAATCATCCGACAAGTTACCGCCCGATTAGCCTCCTCATGTCTCTAGGCAAACTGTATGAGTGTCTGCTCTACAAACGCCTCAGAGACTTCATCTCATCCAAGGGCATTCTCATCGATGAACAATTCGGATTCCGTACAAACCACTCATGCGTTCAACAGGTGCACCGCCTCACGGAGCACATTCTTGTGGGGCTTAATCGACCAAAACCGTTATACACGGGAGCTCTCTTCTTCGACGTCGCAAAAGCGTTCGACAAAGTCTGGCACAACGGTTTGATTTTCAAACTATTCAACATGGGCGTGCCGGATAGTCTCGTGCTCATCATACGGGACTTCTTGTCGAACCGCTCTTTTCGATATCGAGTCGAGGGAACCCGCTCCTCCCCACGACCTCTCACAGCTGGAGTCCCGCAAGGCTCTGTCCTCTCACCCCTCCTATTTAGCTTATTCGTTAACGATATTCCCCGGTCGCCGCCGACCCATTTAGCTTTATTCGCCGACGACACGACTGTTTACTATTCCAGTAGAAATAAGTCCTTAATCGCGAAGAAGCTTCAGAGCGCAGCCCTAGCCCTAGGACAGTGGTTCCGAAAATGGCGCATAGACATCAACCCAGCGAAAAGTACTGCGGTGCTATTTCAGAGGGGAAGCTCCTCACGGATTTCCTCCCGTATTAGGAGAAGGAATCTCACACCCCCGATTACTCTCTTTACACAATCCATACCCTGGGCCAGGAAGGTCAAGTACCTGGGCGTTACCCTGGATGCATCGATGACATTCCGCCCGCATATAAAATCAGTCCGTGACCGTGCCGCGTTTATTCTCGGTAGACTCTACCTCATGATCTGTAAGCGGAGTAAAATGTCCCTTCGGAACAAGGTGACACTTTACAAAACTTGCATAAGGCCCGTCATGACTTACGCGAGTGTGGTGTTCGCTCACGCGGCCCGCACACACATAGACACCCTCCAATCTCTACAATCCCGCTTTTGCAGGTTAGCCGTCGGAGCTTCGTGGTTCGTGAGGAACGTTGACCTACACGACGACCTGGGCCTCGAATCTATCCAGAAATACATGAAGTCAGCGTCGGAACGGTACTTCGATAAGGCTATGCGTCATGATAATCGCCTTATCGTTGCCGCCGCTGACTACTCCCCGAATCCTGATCACGCAGGAGCCAGTCACCGTCGACGCCCTAGACACGTCCTTACGGATCCATCAGATCCAATAACCTTTGCATTAGACGCCTTCAGCTCTAACACTAGGAGCAGGCTTAGGGACTTCGGCTTGAAGTGCAAGCGAGAGCGCGCAACGAGCGACAAAGAGGCACAATCGGCCTCCGCGTTCGGCAGCTTTCGACATCTGTCTCTCTCCTACTTGAGTGAGCGATGCATCCGCGTGGACAGCTGCTATACAACAATACATTTACATGTTTTCGTCAAGTATGAAGTTCAGTGA
Protein
MDAVFAEFLRLRYPKLTSEFQAFKADHTASPLVDSTELAAPASPVPACKASASSTAASVVPAVPASPILASKTAASSAVVTVPAARSSAASVAPSKTPARRSPAPASSSSDSDSEMEVDLAPASSTDEFTLVQKGKKRAAESRAPAAAKISKAANASRPRPQTPVAPPARATPSPRPVAQKKTQTPPPVILQEKAAWDRVSLALKANNINFTNARNLANGIQIKVRTPDDHRALSSYLRKERISFHTYTLQEERELNAVIRGIPKELDAELVKADLLEQGLPVNSVHRMHTGRGREPYNXFLNAYGLANQRDQVSDFLRDHQIDIFLVQETLLKPARRDPKIANYNMVRNNRLSARGGTVIYYRRALHCVPLDPPALEASVCRISLTGHAPIVIASVYLPPDKIVLSSDIEALLGMGSSVILAGDLNCKHIRWNSHTTTPNGRRLDALVDDLAFDIVAPLTPTHYPLNIAHRPDILDIALLKNVTLRLHSIEVVSELDSDHRPVVMKLGRAPDSVPVTRTVVDWHTQGISLAESDPPSLPFSPDSTPSPQDTAEAIDILTSHITSTLDRSSKQVVAEDFLLRFKLSDDIGNSLELKTPRYAPTTGRLAAFDDDEKAELLADTLQTQCTPSTQSVDPVHVELVDSEVERRASLPPSDALPPVTPMEVKDLIKDLRPRKAPGSNGISNRVIKLLPVQVIVMLASIFNAAMTNCIFPAVWKEADVIGIHKPGKPKNHPTSYRPISLLMSLGKLYECLLYKRLRDFISSKGILIDEQFGFRTNHSCVQQVHRLTEHILVGLNRPKPLYTGALFFDVAKAFDKVWHNGLIFKLFNMGVPDSLVLIIRDFLSNRSFRYRVEGTRSSPRPLTAGVPQGSVLSPLLFSLFVNDIPRSPPTHLALFADDTTVYYSSRNKSLIAKKLQSAALALGQWFRKWRIDINPAKSTAVLFQRGSSSRISSRIRRRNLTPPITLFTQSIPWARKVKYLGVTLDASMTFRPHIKSVRDRAAFILGRLYLMICKRSKMSLRNKVTLYKTCIRPVMTYASVVFAHAARTHIDTLQSLQSRFCRLAVGASWFVRNVDLHDDLGLESIQKYMKSASERYFDKAMRHDNRLIVAAADYSPNPDHAGASHRRRPRHVLTDPSDPITFALDAFSSNTRSRLRDFGLKCKRERATSDKEAQSASAFGSFRHLSLSYLSERCIRVDSCYTTIHLHVFVKYEVQ
Summary
Uniprot
EMBL
PRIDE
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
Ontologies
GO
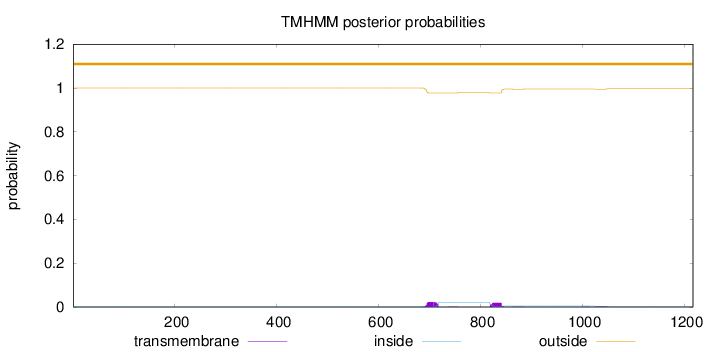
Topology
Length:
1217
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.00327
Exp number, first 60 AAs:
0.00013
Total prob of N-in:
0.00009
outside
1 - 1217
Population Genetic Test Statistics
Pi
119.729868
Theta
30.591279
Tajima's D
2.541866
CLR
0.424535
CSRT
0.951702414879256
Interpretation
Possibly Balancing Selection
