Gene
KWMTBOMO02797
Pre Gene Modal
BGIBMGA009412
Annotation
PREDICTED:_uncharacterized_protein_LOC106716651_[Papilio_machaon]
Transcription factor
Location in the cell
Mitochondrial Reliability : 4.038
Sequence
CDS
ATGCTGTTACGTCCCTTCCATACATTTCCGAATCCACAAAGTTTGATAATAGAACAAATTGAGAGATTTAATAAATGGAAATCTGTACTAAGTGCAGAATCTCAGGAAAAGGGTGAAAAATACATATATAACAACATTCGTCTTTGCAACAGACATTTTGAAAAGTGCTTCATACTACCATCAAATAGACTTACTCAAAATTCAGTCCCAACATTATTTCTTGGACTAAATTATGCAGCTCACATCGAAATTGATCAGCCACAGTTGTTGAGTGGAATTGTGCCGCCTTCAAGTATGAACAATATGATAAAACTTGAAAGTGTATCATACAGAACCTTGAAGAAGAGCTCAAATTCTAATTTGACATATTTGAAATTGATGTCACAAGTAAAAGTGTTAAAGGATAAATGTAGAAAGCAAAAAGATGATATTAAAATGGCAAAGAAGCTTTCCAGGACTAAAGTTTTTCTTAAATACATAGATAAATTACCTGAACCTATAAATCTTTTTACAAAGATGCAATTAAAAAATGCATCTAAGCCTAGAGGGAGGAGATTTTCTCTAAATGAGAAAATTATGTCATTAACTATTCTAAAGCAGAGTCCGAAGGCATATAATTATTTAAGACAGATGTTTGTTTTACCATCAAAACGAAGCCTTCAGACTTTGCTTAGTTTATTTTCTGTCAAACCGGGTTTGAATCTACACATTTTAAAAGATTTACAATCTGCAGTGAGGCTAATGCCTGACGAGAAAAGAATTGTGAATCTAATGTTTGATGAAATATCACTTTGTCCTGGAGTGACCTATGATCCTGATATAGATGCAGTGATAGGATTTGAGGATATGGGATCTAATAGTAGGAATAAAACTCTGGCTGACCATGCACTTGTATTTATGATTAAGAGTGTCAAAAGTAAAAGCAAACAACCCATTTGTTTTACTTTTTGTAAAAGTTCAACAAAAGCTATACACCTAAAAAACCTCATAAAAAATATTGTGCTAGAATTACAAAACATTGGTTTTATTGTTATTACAGTTATTTGCGATCAAGCCAGGACAAATGTCAACGCACTAAAGAACTTACATCAGGATACAAGGGAGAAATATTTGAAGGAAGGACATGGAGATTACATAGAAAAAGCCATTGAAATTGGCGGAAAAAAAATATTTCCTCTATTTGATCCACCACATTTATTGAAAGGTGTGAGAAATAATTTACTGATCAAAGATGCTTTATTTGTGCAGGATGGTGTCATGAAGCGAGCAAAATGGGATCATATTAGAATGTTGTTGGATGTGGATGATGGTGATGACGAAATTAGATTAGTCAATAAACTGACGGAAGCACATGTTATTAAAGAAAAGATCCCAAAAATGAAAGTGAAATTTGCTGCACAAATATTCAGTCAAAGAGTCTCATCAGCTATGAGATTCCTTGCAAGTGAGTGTTGTATATGA
Protein
MLLRPFHTFPNPQSLIIEQIERFNKWKSVLSAESQEKGEKYIYNNIRLCNRHFEKCFILPSNRLTQNSVPTLFLGLNYAAHIEIDQPQLLSGIVPPSSMNNMIKLESVSYRTLKKSSNSNLTYLKLMSQVKVLKDKCRKQKDDIKMAKKLSRTKVFLKYIDKLPEPINLFTKMQLKNASKPRGRRFSLNEKIMSLTILKQSPKAYNYLRQMFVLPSKRSLQTLLSLFSVKPGLNLHILKDLQSAVRLMPDEKRIVNLMFDEISLCPGVTYDPDIDAVIGFEDMGSNSRNKTLADHALVFMIKSVKSKSKQPICFTFCKSSTKAIHLKNLIKNIVLELQNIGFIVITVICDQARTNVNALKNLHQDTREKYLKEGHGDYIEKAIEIGGKKIFPLFDPPHLLKGVRNNLLIKDALFVQDGVMKRAKWDHIRMLLDVDDGDDEIRLVNKLTEAHVIKEKIPKMKVKFAAQIFSQRVSSAMRFLASECCI
Summary
Uniprot
H9JIR1
A0A194QQL0
A0A0N1IC55
A0A2H1VJ77
A0A2A4JPP6
X1XB84
+ More
A0A2A4JYS8 A0A2A4JY71 J9KSA4 J9M961 A0A2A4JZ58 J9M4P5 J9LEP6 A0A2S2NC74 J9LX62 J9LY75 A0A0L7K5T4 X1XC48 A0A2S2P628 J9KEF2 J9KEP3 J9L7T1 A0A2H8TYS2 A0A1Y1NI49 J9M9N1 A0A1Y1LY36 A0A1Y1K6U6 X1XAX4 J9KX16 A0A2S2P5T9 A0A139W9L5 J9LQD8 D6X0C0 J9L5B6 A0A2S2P9C5 A0A139W8P5 J9KFX0 A0A1Y1KFJ5 A0A1Y1KI01 A0A1Y1KN14 J9L862 E9J9D7 A0A1Y1NHP2 X1X0R9 A0A1Y1NHU8 J9KGM8 J9LCP7 J9KCB1 D7EKX8 A0A1Y1KTH5 A0A087SV38
A0A2A4JYS8 A0A2A4JY71 J9KSA4 J9M961 A0A2A4JZ58 J9M4P5 J9LEP6 A0A2S2NC74 J9LX62 J9LY75 A0A0L7K5T4 X1XC48 A0A2S2P628 J9KEF2 J9KEP3 J9L7T1 A0A2H8TYS2 A0A1Y1NI49 J9M9N1 A0A1Y1LY36 A0A1Y1K6U6 X1XAX4 J9KX16 A0A2S2P5T9 A0A139W9L5 J9LQD8 D6X0C0 J9L5B6 A0A2S2P9C5 A0A139W8P5 J9KFX0 A0A1Y1KFJ5 A0A1Y1KI01 A0A1Y1KN14 J9L862 E9J9D7 A0A1Y1NHP2 X1X0R9 A0A1Y1NHU8 J9KGM8 J9LCP7 J9KCB1 D7EKX8 A0A1Y1KTH5 A0A087SV38
EMBL
BABH01032897
KQ461175
KPJ07793.1
KQ461059
KPJ09927.1
ODYU01002680
+ More
SOQ40482.1 NWSH01000959 PCG73380.1 ABLF02011758 ABLF02011759 NWSH01000363 PCG76969.1 PCG76967.1 ABLF02016553 ABLF02016555 ABLF02049531 ABLF02011948 PCG76968.1 ABLF02008684 ABLF02008686 ABLF02008687 ABLF02010326 ABLF02010327 GGMR01002126 MBY14745.1 ABLF02019656 ABLF02019662 ABLF02041486 ABLF02012658 JTDY01010331 KOB58244.1 ABLF02005638 ABLF02005639 GGMR01012308 MBY24927.1 ABLF02028679 ABLF02056730 ABLF02017444 ABLF02017696 GFXV01007611 MBW19416.1 GEZM01001796 JAV97521.1 ABLF02017692 ABLF02017693 GEZM01045715 JAV77678.1 GEZM01090725 JAV57179.1 ABLF02054892 ABLF02023911 GGMR01011939 MBY24558.1 KQ972202 KYB24605.1 ABLF02008211 ABLF02008212 ABLF02008213 ABLF02011424 ABLF02042790 KQ971372 EFA10524.2 ABLF02007924 GGMR01012897 MBY25516.1 KQ973114 KXZ75664.1 ABLF02014046 ABLF02014054 ABLF02014055 ABLF02015166 ABLF02066528 GEZM01085093 JAV60239.1 GEZM01085094 JAV60238.1 GEZM01085095 JAV60237.1 ABLF02003317 GL769271 EFZ10565.1 GEZM01001982 JAV97423.1 ABLF02028653 ABLF02048521 ABLF02061836 ABLF02064667 GEZM01001983 JAV97421.1 ABLF02012220 ABLF02012222 ABLF02053218 ABLF02009194 ABLF02014670 DS497781 EFA11724.1 GEZM01074010 JAV64699.1 KK112098 KFM56727.1
SOQ40482.1 NWSH01000959 PCG73380.1 ABLF02011758 ABLF02011759 NWSH01000363 PCG76969.1 PCG76967.1 ABLF02016553 ABLF02016555 ABLF02049531 ABLF02011948 PCG76968.1 ABLF02008684 ABLF02008686 ABLF02008687 ABLF02010326 ABLF02010327 GGMR01002126 MBY14745.1 ABLF02019656 ABLF02019662 ABLF02041486 ABLF02012658 JTDY01010331 KOB58244.1 ABLF02005638 ABLF02005639 GGMR01012308 MBY24927.1 ABLF02028679 ABLF02056730 ABLF02017444 ABLF02017696 GFXV01007611 MBW19416.1 GEZM01001796 JAV97521.1 ABLF02017692 ABLF02017693 GEZM01045715 JAV77678.1 GEZM01090725 JAV57179.1 ABLF02054892 ABLF02023911 GGMR01011939 MBY24558.1 KQ972202 KYB24605.1 ABLF02008211 ABLF02008212 ABLF02008213 ABLF02011424 ABLF02042790 KQ971372 EFA10524.2 ABLF02007924 GGMR01012897 MBY25516.1 KQ973114 KXZ75664.1 ABLF02014046 ABLF02014054 ABLF02014055 ABLF02015166 ABLF02066528 GEZM01085093 JAV60239.1 GEZM01085094 JAV60238.1 GEZM01085095 JAV60237.1 ABLF02003317 GL769271 EFZ10565.1 GEZM01001982 JAV97423.1 ABLF02028653 ABLF02048521 ABLF02061836 ABLF02064667 GEZM01001983 JAV97421.1 ABLF02012220 ABLF02012222 ABLF02053218 ABLF02009194 ABLF02014670 DS497781 EFA11724.1 GEZM01074010 JAV64699.1 KK112098 KFM56727.1
Proteomes
PRIDE
Pfam
Interpro
IPR021896
Transposase_37
+ More
IPR038441 THAP_Znf_sf
IPR006612 THAP_Znf
IPR008906 HATC_C_dom
IPR025398 DUF4371
IPR003593 AAA+_ATPase
IPR011527 ABC1_TM_dom
IPR027417 P-loop_NTPase
IPR036640 ABC1_TM_sf
IPR030244 Yor1
IPR003439 ABC_transporter-like
IPR017871 ABC_transporter_CS
IPR006580 Znf_TTF
IPR012337 RNaseH-like_sf
IPR023378 YheA/YmcA-like_dom_sf
IPR036361 SAP_dom_sf
IPR003034 SAP_dom
IPR038441 THAP_Znf_sf
IPR006612 THAP_Znf
IPR008906 HATC_C_dom
IPR025398 DUF4371
IPR003593 AAA+_ATPase
IPR011527 ABC1_TM_dom
IPR027417 P-loop_NTPase
IPR036640 ABC1_TM_sf
IPR030244 Yor1
IPR003439 ABC_transporter-like
IPR017871 ABC_transporter_CS
IPR006580 Znf_TTF
IPR012337 RNaseH-like_sf
IPR023378 YheA/YmcA-like_dom_sf
IPR036361 SAP_dom_sf
IPR003034 SAP_dom
Gene 3D
ProteinModelPortal
H9JIR1
A0A194QQL0
A0A0N1IC55
A0A2H1VJ77
A0A2A4JPP6
X1XB84
+ More
A0A2A4JYS8 A0A2A4JY71 J9KSA4 J9M961 A0A2A4JZ58 J9M4P5 J9LEP6 A0A2S2NC74 J9LX62 J9LY75 A0A0L7K5T4 X1XC48 A0A2S2P628 J9KEF2 J9KEP3 J9L7T1 A0A2H8TYS2 A0A1Y1NI49 J9M9N1 A0A1Y1LY36 A0A1Y1K6U6 X1XAX4 J9KX16 A0A2S2P5T9 A0A139W9L5 J9LQD8 D6X0C0 J9L5B6 A0A2S2P9C5 A0A139W8P5 J9KFX0 A0A1Y1KFJ5 A0A1Y1KI01 A0A1Y1KN14 J9L862 E9J9D7 A0A1Y1NHP2 X1X0R9 A0A1Y1NHU8 J9KGM8 J9LCP7 J9KCB1 D7EKX8 A0A1Y1KTH5 A0A087SV38
A0A2A4JYS8 A0A2A4JY71 J9KSA4 J9M961 A0A2A4JZ58 J9M4P5 J9LEP6 A0A2S2NC74 J9LX62 J9LY75 A0A0L7K5T4 X1XC48 A0A2S2P628 J9KEF2 J9KEP3 J9L7T1 A0A2H8TYS2 A0A1Y1NI49 J9M9N1 A0A1Y1LY36 A0A1Y1K6U6 X1XAX4 J9KX16 A0A2S2P5T9 A0A139W9L5 J9LQD8 D6X0C0 J9L5B6 A0A2S2P9C5 A0A139W8P5 J9KFX0 A0A1Y1KFJ5 A0A1Y1KI01 A0A1Y1KN14 J9L862 E9J9D7 A0A1Y1NHP2 X1X0R9 A0A1Y1NHU8 J9KGM8 J9LCP7 J9KCB1 D7EKX8 A0A1Y1KTH5 A0A087SV38
Ontologies
GO
PANTHER
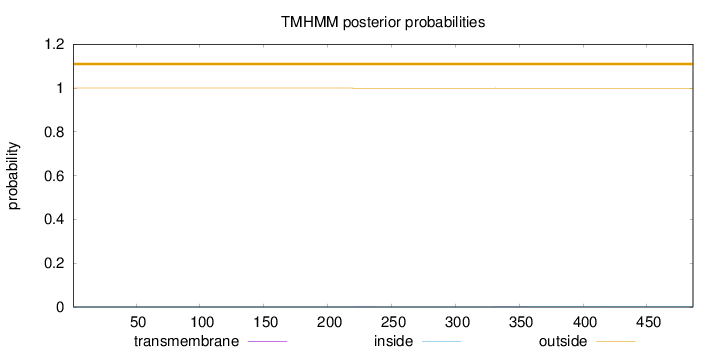
Topology
Length:
486
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01593
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00032
outside
1 - 486
Population Genetic Test Statistics
Pi
224.913976
Theta
188.127368
Tajima's D
0.811294
CLR
0.536727
CSRT
0.601369931503425
Interpretation
Uncertain
