Pre Gene Modal
BGIBMGA003718
Annotation
PREDICTED:_translocating_chain-associated_membrane_protein_1_[Amyelois_transitella]
Full name
Translocating chain-associated membrane protein
Transcription factor
Location in the cell
PlasmaMembrane Reliability : 4.54
Sequence
CDS
ATGGGTGTAAAGCCTGCTATTGGACGAAAAACTAACAAAAACCCTCCCATCTTCAGTCATGAATTTGTAATTCAAAATCATGCAGATATTGTTTCATGTGTTGTTATGGTTTTCCTGGTTGGATTGATGGTACAGTCTACAAGTCCCATTGCAAGCTTGTTCATCAGTTTACATCACAATGTCAGTGGAGTGGAACCAACGAGAGAAGCGCCGAAAGGCGAACCTTTTACTTATGAATCTGGGTGGAAAGATGCTTGTGCCGTGTTTTTCTATTCACTTGTCTGTATTGTGATGCATGCTATTCTTCAGGAGTACTTCCTTGATAAAATTTCCAAGAAGTTCCACCTCTCAAAATCAAGGTTAAGTGCTCTCAATGAATCTGGACAACTTATAGTATTCCATCTCGTAACTCTGGTATGGGGTGGTGATGCTATACTCCGGGAAGGTTTCATTTTCAACATCTCTCAGCTTTGGGAAGGCTACCCAGTGCACCCAATGAGCTTCCTTCTCAAGTTATGGTGGGTTGTCCAAGCTTCCTATTGGTTGCACACAATCCCCGAACTGTACTTCCAACGTATAAAGAAAGATGAGTGGGCAGGACGCATACGAAATGCTGCTACAGCTTTTGGTTTTGTAGCATTGGCTTATGGATTTAATTTCCAACGCGTGGGAGTGTGCCTAGTTGTTCTTCATGCCCTGGCAGAATTTGTGGCACATTGCTATCGTCTCAACACAATCCTCAGGGGAGAACGTGAAGATTTCTTGGACAAGTTTCTAAGTCTTGTAAACGGGACAGTGTTCGTGATCGTTCGCTTGTTCTCTCTCGTCCTTGGAGTGCTTACTTTCTACTTCGGACTCGCCGGTGTCGCACCCTTACTGGTCAGAGTGGCTGCTCTGTCCTTGCTTGTCTCTTTCCAGGTCTACTTGATGTTTAACTTCATATCTGAAGCTATTAAGCAGAGGCAAGAAGCTCGTCAGCAGGCTCTCGCGCGACCTCTCAAGAAAGAGAAGAAGGAAAAACCCAAAAAGGAAAAGGTGAAAAAAGCTCCCGTCGACGAGTCTGACCTGCCCGAGGTGGATCAGAACACGAACAAGACCCTGAGGCAGCGACAGAACGCGACCGCTAAAGCGAAGTGA
Protein
MGVKPAIGRKTNKNPPIFSHEFVIQNHADIVSCVVMVFLVGLMVQSTSPIASLFISLHHNVSGVEPTREAPKGEPFTYESGWKDACAVFFYSLVCIVMHAILQEYFLDKISKKFHLSKSRLSALNESGQLIVFHLVTLVWGGDAILREGFIFNISQLWEGYPVHPMSFLLKLWWVVQASYWLHTIPELYFQRIKKDEWAGRIRNAATAFGFVALAYGFNFQRVGVCLVVLHALAEFVAHCYRLNTILRGEREDFLDKFLSLVNGTVFVIVRLFSLVLGVLTFYFGLAGVAPLLVRVAALSLLVSFQVYLMFNFISEAIKQRQEARQQALARPLKKEKKEKPKKEKVKKAPVDESDLPEVDQNTNKTLRQRQNATAKAK
Summary
Similarity
Belongs to the TRAM family.
Uniprot
H9J2I1
A0A1E1VYA7
A0A2H1W945
A0A2A4K038
S4PMB1
A0A194RGP2
+ More
A0A194PNH1 I4DMD6 A0A212FIW6 A0A0L7LF08 A0A1Q3FCN0 A0A1Q3FCR6 B0W8F6 A0A182HEC5 T1D460 B4JKV7 Q9W5C2 Q1HRQ8 D3TMN1 B4R2K4 B4I980 U5EL95 B4MD36 Q9U1L3 B3P9C1 A0A1W4VFG8 W8B8S1 B3MYD3 A0A1L8EFC1 A0A1B6GYL8 A0A1L8DUN4 T1PBH6 B4L3R6 A0A0K8TPL5 A0A0L0BWV7 A0A336LYK3 A0A336ME24 A0A1A9W8R8 A0A3B0K4J4 A0A1B0DJR7 A0A1I8PHX3 A0A1B6G4W2 A0A0M4EQE8 A0A182NVB6 A0A1B0BNV5 Q29FN4 B4GTT6 A0A1A9XNK5 A0A182F429 A0A2M4A0T8 A0A182YE61 A0A2M4BR54 W5JSJ2 A0A2M3YZ95 A0A0A1X5B0 A0A1A9Z6B9 A0A1B6MD38 A0A182IS04 A0A182Q1V5 Q7QDS0 T1E886 A0A182XA65 A0A182LN48 A0A182HX44 A0A182W8A7 A0A1I8JUJ7 B4ND58 A0A182TN47 A0A067RFZ3 A0A1B6I607 A0A0K8VI85 A0A182UXW6 A0A1B0FHK9 A0A034WL29 A0A1B6M6F2 A0A1J1IIZ6 A0A2R7W525 R4WSP0 A0A2J7QF41 A0A2P8ZEY3 A0A0P6GCW7 A0A0P6IIP4 E9H3M1 A0A0P6ACZ2 T1IL87 A0A0P5ZJ58 A0A1Q3FCT1 A0A1Q3FCW1 A0A1W7R9H0 A0A1E1X8Z1 A0A2I9LPV4 A0A0A9YUL5 A0A0P5VGG1 A0A0P5YN84 A0A0A9YM22 A0A154NZU2 A0A0P5U6B2 E1ZWE7 A0A1E1XUQ5
A0A194PNH1 I4DMD6 A0A212FIW6 A0A0L7LF08 A0A1Q3FCN0 A0A1Q3FCR6 B0W8F6 A0A182HEC5 T1D460 B4JKV7 Q9W5C2 Q1HRQ8 D3TMN1 B4R2K4 B4I980 U5EL95 B4MD36 Q9U1L3 B3P9C1 A0A1W4VFG8 W8B8S1 B3MYD3 A0A1L8EFC1 A0A1B6GYL8 A0A1L8DUN4 T1PBH6 B4L3R6 A0A0K8TPL5 A0A0L0BWV7 A0A336LYK3 A0A336ME24 A0A1A9W8R8 A0A3B0K4J4 A0A1B0DJR7 A0A1I8PHX3 A0A1B6G4W2 A0A0M4EQE8 A0A182NVB6 A0A1B0BNV5 Q29FN4 B4GTT6 A0A1A9XNK5 A0A182F429 A0A2M4A0T8 A0A182YE61 A0A2M4BR54 W5JSJ2 A0A2M3YZ95 A0A0A1X5B0 A0A1A9Z6B9 A0A1B6MD38 A0A182IS04 A0A182Q1V5 Q7QDS0 T1E886 A0A182XA65 A0A182LN48 A0A182HX44 A0A182W8A7 A0A1I8JUJ7 B4ND58 A0A182TN47 A0A067RFZ3 A0A1B6I607 A0A0K8VI85 A0A182UXW6 A0A1B0FHK9 A0A034WL29 A0A1B6M6F2 A0A1J1IIZ6 A0A2R7W525 R4WSP0 A0A2J7QF41 A0A2P8ZEY3 A0A0P6GCW7 A0A0P6IIP4 E9H3M1 A0A0P6ACZ2 T1IL87 A0A0P5ZJ58 A0A1Q3FCT1 A0A1Q3FCW1 A0A1W7R9H0 A0A1E1X8Z1 A0A2I9LPV4 A0A0A9YUL5 A0A0P5VGG1 A0A0P5YN84 A0A0A9YM22 A0A154NZU2 A0A0P5U6B2 E1ZWE7 A0A1E1XUQ5
Pubmed
19121390
23622113
26354079
22651552
22118469
26227816
+ More
26483478 24330624 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17204158 17510324 20353571 24495485 25315136 18057021 26369729 26108605 15632085 25244985 20920257 23761445 25830018 12364791 14747013 17210077 20966253 24845553 25348373 23691247 29403074 21292972 28503490 29248469 25401762 26823975 20798317 29209593
26483478 24330624 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17204158 17510324 20353571 24495485 25315136 18057021 26369729 26108605 15632085 25244985 20920257 23761445 25830018 12364791 14747013 17210077 20966253 24845553 25348373 23691247 29403074 21292972 28503490 29248469 25401762 26823975 20798317 29209593
EMBL
BABH01007979
GDQN01011382
JAT79672.1
ODYU01007126
SOQ49625.1
NWSH01000300
+ More
PCG77631.1 GAIX01003660 JAA88900.1 KQ460207 KPJ16757.1 KQ459597 KPI94867.1 AK402454 BAM19076.1 AGBW02008321 OWR53676.1 JTDY01001343 KOB74138.1 GFDL01009728 JAV25317.1 GFDL01009757 JAV25288.1 DS231858 EDS38885.1 JXUM01035264 JXUM01035265 JXUM01035266 JXUM01035267 JXUM01035268 JXUM01035269 KQ561053 KXJ79861.1 GALA01001088 JAA93764.1 CH916370 EDW00210.1 AE014298 AY075393 AAF45569.1 AAL68227.1 DQ440036 CH477268 ABF18069.1 EAT45439.1 EZ422683 ADD18959.1 CM000366 EDX16804.1 CH480825 EDW43761.1 GANO01001501 JAB58370.1 CH940660 EDW58108.1 AL109630 CAB65875.1 CH954183 EDV45417.1 GAMC01016979 JAB89576.1 CH902632 EDV32627.2 GFDG01001389 JAV17410.1 GECZ01002255 JAS67514.1 GFDF01003947 JAV10137.1 KA646121 AFP60750.1 CH933810 EDW07194.1 GDAI01001522 JAI16081.1 JRES01001224 KNC24491.1 UFQT01000229 SSX22091.1 UFQS01001037 UFQT01001037 SSX08679.1 SSX28594.1 OUUW01000018 SPP89124.1 AJVK01001037 GECZ01012348 JAS57421.1 CP012528 ALC48152.1 JXJN01017617 JXJN01017618 CH379065 EAL31545.1 CH479190 EDW25956.1 GGFK01001105 MBW34426.1 GGFJ01006398 MBW55539.1 ADMH02000590 ETN65729.1 GGFM01000839 MBW21590.1 GBXI01008206 JAD06086.1 GEBQ01006130 JAT33847.1 AXCN02002291 AAAB01008849 EAA07097.3 GAMD01002392 JAA99198.1 APCN01005128 CH964239 EDW82767.1 KK852528 KDR21973.1 GECU01025348 JAS82358.1 GDHF01014084 JAI38230.1 CCAG010014883 GAKP01003593 JAC55359.1 GEBQ01008468 JAT31509.1 CVRI01000048 CRK99038.1 KK854225 PTY13315.1 AK417727 BAN20942.1 NEVH01015301 PNF27221.1 PYGN01000077 PSN55048.1 GDIQ01035088 LRGB01000763 JAN59649.1 KZS16088.1 GDIQ01004033 JAN90704.1 GL732589 EFX73674.1 GDIP01031014 JAM72701.1 JH430751 GDIP01042811 JAM60904.1 GFDL01009695 JAV25350.1 GFDL01009696 JAV25349.1 GFAH01000600 JAV47789.1 GFAC01003712 JAT95476.1 GFWZ01000420 MBW20410.1 GBHO01010344 GDHC01016521 JAG33260.1 JAQ02108.1 GDIP01100596 JAM03119.1 GDIP01056272 JAM47443.1 GBHO01010345 GBRD01007298 GDHC01005997 JAG33259.1 JAG58523.1 JAQ12632.1 KQ434786 KZC05102.1 GDIP01121253 JAL82461.1 GL434819 EFN74493.1 GFAA01000578 JAU02857.1
PCG77631.1 GAIX01003660 JAA88900.1 KQ460207 KPJ16757.1 KQ459597 KPI94867.1 AK402454 BAM19076.1 AGBW02008321 OWR53676.1 JTDY01001343 KOB74138.1 GFDL01009728 JAV25317.1 GFDL01009757 JAV25288.1 DS231858 EDS38885.1 JXUM01035264 JXUM01035265 JXUM01035266 JXUM01035267 JXUM01035268 JXUM01035269 KQ561053 KXJ79861.1 GALA01001088 JAA93764.1 CH916370 EDW00210.1 AE014298 AY075393 AAF45569.1 AAL68227.1 DQ440036 CH477268 ABF18069.1 EAT45439.1 EZ422683 ADD18959.1 CM000366 EDX16804.1 CH480825 EDW43761.1 GANO01001501 JAB58370.1 CH940660 EDW58108.1 AL109630 CAB65875.1 CH954183 EDV45417.1 GAMC01016979 JAB89576.1 CH902632 EDV32627.2 GFDG01001389 JAV17410.1 GECZ01002255 JAS67514.1 GFDF01003947 JAV10137.1 KA646121 AFP60750.1 CH933810 EDW07194.1 GDAI01001522 JAI16081.1 JRES01001224 KNC24491.1 UFQT01000229 SSX22091.1 UFQS01001037 UFQT01001037 SSX08679.1 SSX28594.1 OUUW01000018 SPP89124.1 AJVK01001037 GECZ01012348 JAS57421.1 CP012528 ALC48152.1 JXJN01017617 JXJN01017618 CH379065 EAL31545.1 CH479190 EDW25956.1 GGFK01001105 MBW34426.1 GGFJ01006398 MBW55539.1 ADMH02000590 ETN65729.1 GGFM01000839 MBW21590.1 GBXI01008206 JAD06086.1 GEBQ01006130 JAT33847.1 AXCN02002291 AAAB01008849 EAA07097.3 GAMD01002392 JAA99198.1 APCN01005128 CH964239 EDW82767.1 KK852528 KDR21973.1 GECU01025348 JAS82358.1 GDHF01014084 JAI38230.1 CCAG010014883 GAKP01003593 JAC55359.1 GEBQ01008468 JAT31509.1 CVRI01000048 CRK99038.1 KK854225 PTY13315.1 AK417727 BAN20942.1 NEVH01015301 PNF27221.1 PYGN01000077 PSN55048.1 GDIQ01035088 LRGB01000763 JAN59649.1 KZS16088.1 GDIQ01004033 JAN90704.1 GL732589 EFX73674.1 GDIP01031014 JAM72701.1 JH430751 GDIP01042811 JAM60904.1 GFDL01009695 JAV25350.1 GFDL01009696 JAV25349.1 GFAH01000600 JAV47789.1 GFAC01003712 JAT95476.1 GFWZ01000420 MBW20410.1 GBHO01010344 GDHC01016521 JAG33260.1 JAQ02108.1 GDIP01100596 JAM03119.1 GDIP01056272 JAM47443.1 GBHO01010345 GBRD01007298 GDHC01005997 JAG33259.1 JAG58523.1 JAQ12632.1 KQ434786 KZC05102.1 GDIP01121253 JAL82461.1 GL434819 EFN74493.1 GFAA01000578 JAU02857.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000002320 UP000069940 UP000249989 UP000001070 UP000000803 UP000008820 UP000000304 UP000001292 UP000008792 UP000008711 UP000192221 UP000007801 UP000095301 UP000009192 UP000037069 UP000091820 UP000268350 UP000092462 UP000095300 UP000092553 UP000075884 UP000092460 UP000001819 UP000008744 UP000092443 UP000069272 UP000076408 UP000000673 UP000092445 UP000075880 UP000075886 UP000007062 UP000076407 UP000075882 UP000075840 UP000075920 UP000075900 UP000007798 UP000075902 UP000027135 UP000075903 UP000092444 UP000183832 UP000235965 UP000245037 UP000076858 UP000000305 UP000076502 UP000000311
UP000002320 UP000069940 UP000249989 UP000001070 UP000000803 UP000008820 UP000000304 UP000001292 UP000008792 UP000008711 UP000192221 UP000007801 UP000095301 UP000009192 UP000037069 UP000091820 UP000268350 UP000092462 UP000095300 UP000092553 UP000075884 UP000092460 UP000001819 UP000008744 UP000092443 UP000069272 UP000076408 UP000000673 UP000092445 UP000075880 UP000075886 UP000007062 UP000076407 UP000075882 UP000075840 UP000075920 UP000075900 UP000007798 UP000075902 UP000027135 UP000075903 UP000092444 UP000183832 UP000235965 UP000245037 UP000076858 UP000000305 UP000076502 UP000000311
Interpro
ProteinModelPortal
H9J2I1
A0A1E1VYA7
A0A2H1W945
A0A2A4K038
S4PMB1
A0A194RGP2
+ More
A0A194PNH1 I4DMD6 A0A212FIW6 A0A0L7LF08 A0A1Q3FCN0 A0A1Q3FCR6 B0W8F6 A0A182HEC5 T1D460 B4JKV7 Q9W5C2 Q1HRQ8 D3TMN1 B4R2K4 B4I980 U5EL95 B4MD36 Q9U1L3 B3P9C1 A0A1W4VFG8 W8B8S1 B3MYD3 A0A1L8EFC1 A0A1B6GYL8 A0A1L8DUN4 T1PBH6 B4L3R6 A0A0K8TPL5 A0A0L0BWV7 A0A336LYK3 A0A336ME24 A0A1A9W8R8 A0A3B0K4J4 A0A1B0DJR7 A0A1I8PHX3 A0A1B6G4W2 A0A0M4EQE8 A0A182NVB6 A0A1B0BNV5 Q29FN4 B4GTT6 A0A1A9XNK5 A0A182F429 A0A2M4A0T8 A0A182YE61 A0A2M4BR54 W5JSJ2 A0A2M3YZ95 A0A0A1X5B0 A0A1A9Z6B9 A0A1B6MD38 A0A182IS04 A0A182Q1V5 Q7QDS0 T1E886 A0A182XA65 A0A182LN48 A0A182HX44 A0A182W8A7 A0A1I8JUJ7 B4ND58 A0A182TN47 A0A067RFZ3 A0A1B6I607 A0A0K8VI85 A0A182UXW6 A0A1B0FHK9 A0A034WL29 A0A1B6M6F2 A0A1J1IIZ6 A0A2R7W525 R4WSP0 A0A2J7QF41 A0A2P8ZEY3 A0A0P6GCW7 A0A0P6IIP4 E9H3M1 A0A0P6ACZ2 T1IL87 A0A0P5ZJ58 A0A1Q3FCT1 A0A1Q3FCW1 A0A1W7R9H0 A0A1E1X8Z1 A0A2I9LPV4 A0A0A9YUL5 A0A0P5VGG1 A0A0P5YN84 A0A0A9YM22 A0A154NZU2 A0A0P5U6B2 E1ZWE7 A0A1E1XUQ5
A0A194PNH1 I4DMD6 A0A212FIW6 A0A0L7LF08 A0A1Q3FCN0 A0A1Q3FCR6 B0W8F6 A0A182HEC5 T1D460 B4JKV7 Q9W5C2 Q1HRQ8 D3TMN1 B4R2K4 B4I980 U5EL95 B4MD36 Q9U1L3 B3P9C1 A0A1W4VFG8 W8B8S1 B3MYD3 A0A1L8EFC1 A0A1B6GYL8 A0A1L8DUN4 T1PBH6 B4L3R6 A0A0K8TPL5 A0A0L0BWV7 A0A336LYK3 A0A336ME24 A0A1A9W8R8 A0A3B0K4J4 A0A1B0DJR7 A0A1I8PHX3 A0A1B6G4W2 A0A0M4EQE8 A0A182NVB6 A0A1B0BNV5 Q29FN4 B4GTT6 A0A1A9XNK5 A0A182F429 A0A2M4A0T8 A0A182YE61 A0A2M4BR54 W5JSJ2 A0A2M3YZ95 A0A0A1X5B0 A0A1A9Z6B9 A0A1B6MD38 A0A182IS04 A0A182Q1V5 Q7QDS0 T1E886 A0A182XA65 A0A182LN48 A0A182HX44 A0A182W8A7 A0A1I8JUJ7 B4ND58 A0A182TN47 A0A067RFZ3 A0A1B6I607 A0A0K8VI85 A0A182UXW6 A0A1B0FHK9 A0A034WL29 A0A1B6M6F2 A0A1J1IIZ6 A0A2R7W525 R4WSP0 A0A2J7QF41 A0A2P8ZEY3 A0A0P6GCW7 A0A0P6IIP4 E9H3M1 A0A0P6ACZ2 T1IL87 A0A0P5ZJ58 A0A1Q3FCT1 A0A1Q3FCW1 A0A1W7R9H0 A0A1E1X8Z1 A0A2I9LPV4 A0A0A9YUL5 A0A0P5VGG1 A0A0P5YN84 A0A0A9YM22 A0A154NZU2 A0A0P5U6B2 E1ZWE7 A0A1E1XUQ5
Ontologies
GO
PANTHER
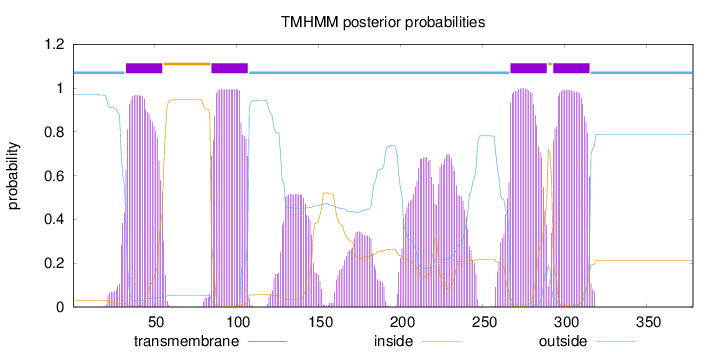
Topology
Length:
378
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
130.71716
Exp number, first 60 AAs:
21.77987
Total prob of N-in:
0.96882
POSSIBLE N-term signal
sequence
inside
1 - 32
TMhelix
33 - 55
outside
56 - 84
TMhelix
85 - 107
inside
108 - 266
TMhelix
267 - 289
outside
290 - 292
TMhelix
293 - 315
inside
316 - 378
Population Genetic Test Statistics
Pi
219.205246
Theta
155.174865
Tajima's D
1.20427
CLR
0.005223
CSRT
0.71966401679916
Interpretation
Uncertain
