Gene
KWMTBOMO01855
Pre Gene Modal
BGIBMGA006184
Annotation
PREDICTED:_protein_ultraspiracle_homolog_isoform_X1_[Bombyx_mori]
Full name
Protein ultraspiracle homolog
Alternative Name
BmCF1
Nuclear receptor subfamily 2 group B member 4
RXR type hormone receptor CF1
Nuclear receptor subfamily 2 group B member 4
RXR type hormone receptor CF1
Transcription factor
Location in the cell
Mitochondrial Reliability : 1.593 Nuclear Reliability : 1.457
Sequence
CDS
ATGACGCCTAGCCCACAGCAGCAGCAGCAGATGGTGCCGTCTACACAGCATTCGAATTTCCTGCAGCCAATGGCTACACCTTCAACCACACCCAATGTTGAACTCGATATACAATGGCTGAACATAGAGTCAGGGTTTATGTCGCCTATGTCACCGCCCGAAATGAAGCCAGATACGGCGATGCTCGACGGTTTCCGAGACGACTCGACGCCGCCCCCTCCCTTCAAGAATTATCCCCCTAACCATCCCTTGAGCGGCTCAAAACATCTATGCTCCATATGTGGCGACAGAGCCTCGGGCAAACATTACGGAGTATACAGTTGTGAAGGCTGCAAAGGGTTCTTCAAGAGGACAGTCAGGAAGGATCTCACATACGCGTGCCGGGAGGACAAGAATTGTATAATAGATAAACGCCAGCGGAATCGTTGCCAGTACTGCCGATACCAGAAGTGTCTCGCTTGCGGCATGAAGAGGGAGGCTGTGCAAGAGGAGAGACAGCGAGCCGCGAGGGGTACAGAAGACGCTCATCCCAGCAGCTCTGTACAGGAGCTATCGATCGAGCGGCTGCTGGAATTGGAGGCGTTAGTTGCGGATTCAGCTGAGGAGTTACAGATCCTACGCGTCGGTCCCGAAAGCGGCGTGCCGGCCAAGTACCGAGCTCCCGTTTCGAGTCTTTGTCAAATAGGCAACAAACAGATAGCCGCTCTCATTGTTTGGGCGCGTGACATTCCACACTTCGGGCAGCTAGAAATCGACGATCAGATCCTTCTAATCAAGGGCTCCTGGAACGAACTGCTGCTGTTCGCTATCGCATGGCGGTCTATGGAGTTCCTCAATGATGAACGAGAGAACGTAGACTCGCGGAATACGGCGCCGCCTCAACTCATTTGTTTAATGCCAGGCATGACGCTGCACCGCAACTCCGCGCTGCAGGCCGGCGTGGGGCAGATCTTCGACCGCGTGCTCTCCGAGCTGTCGCTCAAGATGCGCTCCCTCCGCATGGACCAGGCCGAGTACGTCGCGCTCAAGGCCATCATACTCCTCAATCCTGACGTAAAAGGATTGAAGAATAAACAAGAAGTGGACGTTCTTCGAGAAAAGATGTTCTTATGCCTGGACGAGTACTGCCGGCGCTCGCGCGGCGGGGAGGAGGGTCGGTTCGCGGCGCTGCTGCTGCGGCTGCCGGCGCTGCGCTCCATCTCGCTCAAGAGCTTCGAGCACCTCTACCTGTTCCACCTCGTGGCCGAGGGCAGCGTGAGCTCGTACATCCGCGACGCGCTCTGCAACCACGCGCCGCCCATCGACACCAACATCATGTAG
Protein
MTPSPQQQQQMVPSTQHSNFLQPMATPSTTPNVELDIQWLNIESGFMSPMSPPEMKPDTAMLDGFRDDSTPPPPFKNYPPNHPLSGSKHLCSICGDRASGKHYGVYSCEGCKGFFKRTVRKDLTYACREDKNCIIDKRQRNRCQYCRYQKCLACGMKREAVQEERQRAARGTEDAHPSSSVQELSIERLLELEALVADSAEELQILRVGPESGVPAKYRAPVSSLCQIGNKQIAALIVWARDIPHFGQLEIDDQILLIKGSWNELLLFAIAWRSMEFLNDERENVDSRNTAPPQLICLMPGMTLHRNSALQAGVGQIFDRVLSELSLKMRSLRMDQAEYVALKAIILLNPDVKGLKNKQEVDVLREKMFLCLDEYCRRSRGGEEGRFAALLLRLPALRSISLKSFEHLYLFHLVAEGSVSSYIRDALCNHAPPIDTNIM
Summary
Description
Receptor for ecdysone. May be an important modulator of insect metamorphosis (By similarity).
Subunit
Heterodimer of USP and ECR.
Heterodimer of USP and ECR. Only the heterodimer is capable of high-affinity binding to ecdysone (By similarity).
Heterodimer of USP and ECR. Only the heterodimer is capable of high-affinity binding to ecdysone (By similarity).
Similarity
Belongs to the nuclear hormone receptor family. NR2 subfamily.
Belongs to the nuclear hormone receptor family.
Belongs to the nuclear hormone receptor family.
Keywords
Complete proteome
DNA-binding
Metal-binding
Nucleus
Receptor
Reference proteome
Transcription
Transcription regulation
Zinc
Zinc-finger
Feature
chain Protein ultraspiracle homolog
Uniprot
P49700
P54779
A0A2W1BFV9
A0A1V0J8W3
M4IQQ9
K7WYJ0
+ More
S5YEA6 A9UEF7 A0A2H4Z8R4 E3UJZ6 O76202 A0A2A4K9Z3 A0A194PKF8 B9UCQ5 B2ZCQ5 K7WLB9 A0A2A4K9M0 A0A0M5KTU5 S5XLP8 Q8I1M8 I3RQC0 Q6IVK1 G9M5R1 G5CV12 A0A212F7B4 A0A194R7V9 A0A0L7LGU6 Q1AHP6 A0A1D6YNX1 A0A1L8DHL9 U5EZR1 A0A1L8DHH7 A0A0K8TPF2 A0A336LQ69 A0A336N026 K0BY27 A0A336N4B2 Q9GSG7 A0A1S4EVT0 Q9GSG8 Q0IGE0 A0A1S4EVR8 A0A182HDC9 A0A023EU70 B0X9K6 A0A1Q3FWN1 A0A084VW20 A0A0A1WR56 A0A182MKL1 A0A182RT62 A0A182VRF8 A0A182UL84 A0A182XW12 A0A182PE19 Q9U3Y3 T1DL21 W8C145 F5HN02 E0Z5Q8 A0A182HZX0 A0A182XGF6 F5HN03 A0A0K8VTG0 A0A034VVC4 A0A182LQZ6 A0A182V3C6 A0A182N6B8 A0A182QE69 A0A182IQI7 A0A2M4AKG3 A0A2M4AKL7 A0A2M4AJP8 A0A2M4AJP7 A0A182F802 A0A2M4AJT5 A0A2M4CWG7 W5JAL7 A0A1A9W9F4 A0A1A9XH09 A0A1B0FMJ8 A0A1A9VRP2 A0A1A9Z1M9 A0A1B0AND1 K4MJU9 A0A1I8NWH7 T1P9L7 K4MV69 Q962I5 A0A0L0BR06 A0A1J1HRR1 T1GBH8
S5YEA6 A9UEF7 A0A2H4Z8R4 E3UJZ6 O76202 A0A2A4K9Z3 A0A194PKF8 B9UCQ5 B2ZCQ5 K7WLB9 A0A2A4K9M0 A0A0M5KTU5 S5XLP8 Q8I1M8 I3RQC0 Q6IVK1 G9M5R1 G5CV12 A0A212F7B4 A0A194R7V9 A0A0L7LGU6 Q1AHP6 A0A1D6YNX1 A0A1L8DHL9 U5EZR1 A0A1L8DHH7 A0A0K8TPF2 A0A336LQ69 A0A336N026 K0BY27 A0A336N4B2 Q9GSG7 A0A1S4EVT0 Q9GSG8 Q0IGE0 A0A1S4EVR8 A0A182HDC9 A0A023EU70 B0X9K6 A0A1Q3FWN1 A0A084VW20 A0A0A1WR56 A0A182MKL1 A0A182RT62 A0A182VRF8 A0A182UL84 A0A182XW12 A0A182PE19 Q9U3Y3 T1DL21 W8C145 F5HN02 E0Z5Q8 A0A182HZX0 A0A182XGF6 F5HN03 A0A0K8VTG0 A0A034VVC4 A0A182LQZ6 A0A182V3C6 A0A182N6B8 A0A182QE69 A0A182IQI7 A0A2M4AKG3 A0A2M4AKL7 A0A2M4AJP8 A0A2M4AJP7 A0A182F802 A0A2M4AJT5 A0A2M4CWG7 W5JAL7 A0A1A9W9F4 A0A1A9XH09 A0A1B0FMJ8 A0A1A9VRP2 A0A1A9Z1M9 A0A1B0AND1 K4MJU9 A0A1I8NWH7 T1P9L7 K4MV69 Q962I5 A0A0L0BR06 A0A1J1HRR1 T1GBH8
Pubmed
8176738
9013254
28756777
23523827
22044719
9581288
+ More
26354079 26325588 12459199 15796748 22118469 26227816 16756554 26369729 8892313 9765285 17510324 26483478 24945155 24438588 25830018 25244985 10644973 24495485 12364791 14747013 17210077 25348373 20966253 20920257 23761445 23039844 25315136 11378412 26108605
26354079 26325588 12459199 15796748 22118469 26227816 16756554 26369729 8892313 9765285 17510324 26483478 24945155 24438588 25830018 25244985 10644973 24495485 12364791 14747013 17210077 25348373 20966253 20920257 23761445 23039844 25315136 11378412 26108605
EMBL
U06073
U44837
KZ150176
PZC72564.1
KY411160
ARD05166.1
+ More
JQ731606 AGA17964.1 JX680324 AFX60116.1 KC953864 AGT02382.1 EU180022 JQ730733 ABX79144.1 AFK27932.1 KY488556 AUF71655.1 HM445735 ADO64596.1 AF016368 NWSH01000026 PCG80603.1 KQ459601 KPI93926.1 EU526832 ACD74808.1 EU642475 ACD39740.1 JX680325 AFX60117.1 PCG80604.1 KT343806 ALD83751.1 KC953863 AGT02381.1 AB081840 BAC53670.1 JQ730734 AFK27933.1 AY619987 AAT44330.1 AB568470 BAL41655.1 GU222670 JN410831 ADA61199.1 AEP25407.1 AGBW02009889 OWR49634.1 KQ460855 KPJ11956.1 JTDY01001136 KOB74773.1 DQ083513 AAZ38141.1 KR815909 ANF34832.1 GFDF01008250 JAV05834.1 GANO01000320 JAB59551.1 GFDF01008249 JAV05835.1 GDAI01001582 JAI16021.1 UFQS01000101 UFQT01000101 SSW99595.1 SSX19975.1 UFQS01005117 UFQT01005117 SSX16712.1 SSX35904.1 GQ427084 AFT63499.1 UFQS01005149 UFQT01005149 SSX16732.1 SSX35923.1 AF305214 CH477192 AAG24887.1 EJY57333.1 AF305213 AAG24886.1 EAT48598.1 JXUM01001635 JXUM01001636 KQ560125 KXJ84343.1 GAPW01001121 JAC12477.1 DS232541 EDS43175.1 GFDL01003169 JAV31876.1 ATLV01017380 KE525164 KFB42164.1 GBXI01013384 GBXI01002981 JAD00908.1 JAD11311.1 AXCM01005402 AF210734 AAF19033.1 GAMD01000661 JAB00930.1 GAMC01001143 JAC05413.1 AAAB01008987 EGK97384.1 HM195185 ADM64635.1 APCN01002017 EAA01003.6 EGK97383.1 EGK97385.1 GDHF01010171 JAI42143.1 GAKP01013459 JAC45493.1 AXCN02000281 AXCN02000282 GGFK01007952 MBW41273.1 GGFK01007951 MBW41272.1 GGFK01007688 MBW41009.1 GGFK01007689 MBW41010.1 GGFK01007686 MBW41007.1 GGFL01005504 MBW69682.1 ADMH02002036 ETN59829.1 CCAG010001067 JXJN01000796 JQ679100 AFV25497.1 KA645422 AFP60051.1 JQ679104 AFV25501.1 AY007213 AAG01569.1 JRES01001496 KNC22467.1 CVRI01000015 CRK90076.1 CAQQ02103930
JQ731606 AGA17964.1 JX680324 AFX60116.1 KC953864 AGT02382.1 EU180022 JQ730733 ABX79144.1 AFK27932.1 KY488556 AUF71655.1 HM445735 ADO64596.1 AF016368 NWSH01000026 PCG80603.1 KQ459601 KPI93926.1 EU526832 ACD74808.1 EU642475 ACD39740.1 JX680325 AFX60117.1 PCG80604.1 KT343806 ALD83751.1 KC953863 AGT02381.1 AB081840 BAC53670.1 JQ730734 AFK27933.1 AY619987 AAT44330.1 AB568470 BAL41655.1 GU222670 JN410831 ADA61199.1 AEP25407.1 AGBW02009889 OWR49634.1 KQ460855 KPJ11956.1 JTDY01001136 KOB74773.1 DQ083513 AAZ38141.1 KR815909 ANF34832.1 GFDF01008250 JAV05834.1 GANO01000320 JAB59551.1 GFDF01008249 JAV05835.1 GDAI01001582 JAI16021.1 UFQS01000101 UFQT01000101 SSW99595.1 SSX19975.1 UFQS01005117 UFQT01005117 SSX16712.1 SSX35904.1 GQ427084 AFT63499.1 UFQS01005149 UFQT01005149 SSX16732.1 SSX35923.1 AF305214 CH477192 AAG24887.1 EJY57333.1 AF305213 AAG24886.1 EAT48598.1 JXUM01001635 JXUM01001636 KQ560125 KXJ84343.1 GAPW01001121 JAC12477.1 DS232541 EDS43175.1 GFDL01003169 JAV31876.1 ATLV01017380 KE525164 KFB42164.1 GBXI01013384 GBXI01002981 JAD00908.1 JAD11311.1 AXCM01005402 AF210734 AAF19033.1 GAMD01000661 JAB00930.1 GAMC01001143 JAC05413.1 AAAB01008987 EGK97384.1 HM195185 ADM64635.1 APCN01002017 EAA01003.6 EGK97383.1 EGK97385.1 GDHF01010171 JAI42143.1 GAKP01013459 JAC45493.1 AXCN02000281 AXCN02000282 GGFK01007952 MBW41273.1 GGFK01007951 MBW41272.1 GGFK01007688 MBW41009.1 GGFK01007689 MBW41010.1 GGFK01007686 MBW41007.1 GGFL01005504 MBW69682.1 ADMH02002036 ETN59829.1 CCAG010001067 JXJN01000796 JQ679100 AFV25497.1 KA645422 AFP60051.1 JQ679104 AFV25501.1 AY007213 AAG01569.1 JRES01001496 KNC22467.1 CVRI01000015 CRK90076.1 CAQQ02103930
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000037510
+ More
UP000008820 UP000069940 UP000249989 UP000002320 UP000030765 UP000075883 UP000075900 UP000075920 UP000075902 UP000076408 UP000075885 UP000007062 UP000075840 UP000076407 UP000075882 UP000075903 UP000075884 UP000075886 UP000075880 UP000069272 UP000000673 UP000091820 UP000092443 UP000092444 UP000078200 UP000092445 UP000092460 UP000095300 UP000095301 UP000037069 UP000183832 UP000015102
UP000008820 UP000069940 UP000249989 UP000002320 UP000030765 UP000075883 UP000075900 UP000075920 UP000075902 UP000076408 UP000075885 UP000007062 UP000075840 UP000076407 UP000075882 UP000075903 UP000075884 UP000075886 UP000075880 UP000069272 UP000000673 UP000091820 UP000092443 UP000092444 UP000078200 UP000092445 UP000092460 UP000095300 UP000095301 UP000037069 UP000183832 UP000015102
Interpro
SUPFAM
SSF48508
SSF48508
Gene 3D
ProteinModelPortal
P49700
P54779
A0A2W1BFV9
A0A1V0J8W3
M4IQQ9
K7WYJ0
+ More
S5YEA6 A9UEF7 A0A2H4Z8R4 E3UJZ6 O76202 A0A2A4K9Z3 A0A194PKF8 B9UCQ5 B2ZCQ5 K7WLB9 A0A2A4K9M0 A0A0M5KTU5 S5XLP8 Q8I1M8 I3RQC0 Q6IVK1 G9M5R1 G5CV12 A0A212F7B4 A0A194R7V9 A0A0L7LGU6 Q1AHP6 A0A1D6YNX1 A0A1L8DHL9 U5EZR1 A0A1L8DHH7 A0A0K8TPF2 A0A336LQ69 A0A336N026 K0BY27 A0A336N4B2 Q9GSG7 A0A1S4EVT0 Q9GSG8 Q0IGE0 A0A1S4EVR8 A0A182HDC9 A0A023EU70 B0X9K6 A0A1Q3FWN1 A0A084VW20 A0A0A1WR56 A0A182MKL1 A0A182RT62 A0A182VRF8 A0A182UL84 A0A182XW12 A0A182PE19 Q9U3Y3 T1DL21 W8C145 F5HN02 E0Z5Q8 A0A182HZX0 A0A182XGF6 F5HN03 A0A0K8VTG0 A0A034VVC4 A0A182LQZ6 A0A182V3C6 A0A182N6B8 A0A182QE69 A0A182IQI7 A0A2M4AKG3 A0A2M4AKL7 A0A2M4AJP8 A0A2M4AJP7 A0A182F802 A0A2M4AJT5 A0A2M4CWG7 W5JAL7 A0A1A9W9F4 A0A1A9XH09 A0A1B0FMJ8 A0A1A9VRP2 A0A1A9Z1M9 A0A1B0AND1 K4MJU9 A0A1I8NWH7 T1P9L7 K4MV69 Q962I5 A0A0L0BR06 A0A1J1HRR1 T1GBH8
S5YEA6 A9UEF7 A0A2H4Z8R4 E3UJZ6 O76202 A0A2A4K9Z3 A0A194PKF8 B9UCQ5 B2ZCQ5 K7WLB9 A0A2A4K9M0 A0A0M5KTU5 S5XLP8 Q8I1M8 I3RQC0 Q6IVK1 G9M5R1 G5CV12 A0A212F7B4 A0A194R7V9 A0A0L7LGU6 Q1AHP6 A0A1D6YNX1 A0A1L8DHL9 U5EZR1 A0A1L8DHH7 A0A0K8TPF2 A0A336LQ69 A0A336N026 K0BY27 A0A336N4B2 Q9GSG7 A0A1S4EVT0 Q9GSG8 Q0IGE0 A0A1S4EVR8 A0A182HDC9 A0A023EU70 B0X9K6 A0A1Q3FWN1 A0A084VW20 A0A0A1WR56 A0A182MKL1 A0A182RT62 A0A182VRF8 A0A182UL84 A0A182XW12 A0A182PE19 Q9U3Y3 T1DL21 W8C145 F5HN02 E0Z5Q8 A0A182HZX0 A0A182XGF6 F5HN03 A0A0K8VTG0 A0A034VVC4 A0A182LQZ6 A0A182V3C6 A0A182N6B8 A0A182QE69 A0A182IQI7 A0A2M4AKG3 A0A2M4AKL7 A0A2M4AJP8 A0A2M4AJP7 A0A182F802 A0A2M4AJT5 A0A2M4CWG7 W5JAL7 A0A1A9W9F4 A0A1A9XH09 A0A1B0FMJ8 A0A1A9VRP2 A0A1A9Z1M9 A0A1B0AND1 K4MJU9 A0A1I8NWH7 T1P9L7 K4MV69 Q962I5 A0A0L0BR06 A0A1J1HRR1 T1GBH8
PDB
4UMM
E-value=5.00596e-120,
Score=1104
Ontologies
GO
Topology
Subcellular location
Nucleus
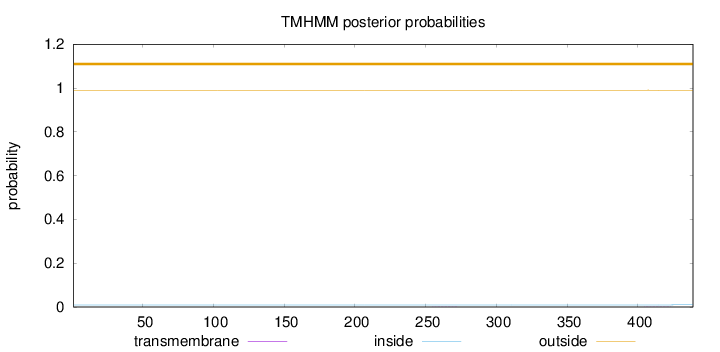
Length:
439
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00869
Exp number, first 60 AAs:
0.00022
Total prob of N-in:
0.00960
outside
1 - 439
Population Genetic Test Statistics
Pi
22.216723
Theta
32.027251
Tajima's D
0.175397
CLR
131.11118
CSRT
0.417829108544573
Interpretation
Uncertain
