Gene
KWMTBOMO01220
Pre Gene Modal
BGIBMGA009541
Annotation
PREDICTED:_protein_odd-skipped_[Bombyx_mori]
Full name
Protein odd-skipped
Transcription factor
Location in the cell
Nuclear Reliability : 4.129
Sequence
CDS
ATGGAAGATAAGCCGTACAGTGTTGATAGTTCGGACGATATTAACGTGGACGACGACGATGAGCATACCGAAGAAAATCACAGCTACCCACCAGACTTCCCTATAGAAGCATTGGTAAAATTAGAGCGACCGTCACCCCCTTTGCCGACGCCACCACACACACCCACAGACGACACGCCCCCAGCTGTCGTTATCTCCCGTCACCAACCCGCGTGGATTCCACCCGCACCCCAATCGTACCCCACCTATCCTCGTCCAATGCAATCTCCGTACTCCTACGATAAAATGCCCTACTCACACCCGGCCATGTCAGCGTACATGCAGGCGCAAGTAGCGACGCTGCCACCCTCCGCCCCATACCACGCTATGCTCGTGAATCAGTGGATAAGAAACGCCGCGCTGTATCATCATTCCCTGCGCTATCCGGGAGTGATGGCCCAAAGAATGCCCGGAAGAAATCCTCCGCCGCTAAGAGCGCCAAGCGTTCCCGGAGCGAGACCAAAGAAACAGTTCATCTGCAAATACTGCAATCGCCAATTCACAAAGTCCTACAACTTATTGATTCACGAGAGAACGCACACTGACGAGAGACCTTACTCGTGCGACATTTGCGGGAAGGCATTCAGAAGACAAGACCACCTGAGAGATCATAGGTACATTCATTCGAAGGAGAAGCCGTTTAAGTGCACGGAATGCGGGAAAGGATTCTGCCAATCGAGAACGTTAGCGGTGCACAAGATTTTGCACATGGAAGAATCTCCGCACAAATGTCCGGTCTGCAACAAGAGCTTCAACCAAAGATCTAATCTGAAGACGCATTTGCTGACGCACAGCGAAGCGAACAAACACCATTTGGACCATTTGGATGGTTGTCAAGAGGTCTCTTCAACAAGCCAAACGGACAGCTCTTTGTTGGATCTGTCCCATAAAAGTCCCGTTGCGTTACCGCAACAGCTTCAGCCACTTGTCAGCCCTCATCAATCTCCTGTCGAAGGGCCGAAAAGGACATTAGGATTCTCAATAGAGGACATTATGAAACGTTAA
Protein
MEDKPYSVDSSDDINVDDDDEHTEENHSYPPDFPIEALVKLERPSPPLPTPPHTPTDDTPPAVVISRHQPAWIPPAPQSYPTYPRPMQSPYSYDKMPYSHPAMSAYMQAQVATLPPSAPYHAMLVNQWIRNAALYHHSLRYPGVMAQRMPGRNPPPLRAPSVPGARPKKQFICKYCNRQFTKSYNLLIHERTHTDERPYSCDICGKAFRRQDHLRDHRYIHSKEKPFKCTECGKGFCQSRTLAVHKILHMEESPHKCPVCNKSFNQRSNLKTHLLTHSEANKHHLDHLDGCQEVSSTSQTDSSLLDLSHKSPVALPQQLQPLVSPHQSPVEGPKRTLGFSIEDIMKR
Summary
Description
Pair-rule protein that determines both the size and polarity of even-numbered as well as odd-numbered parasegments during embryogenesis. DNA-binding transcription factor that acts primarily as a transcriptional repressor but can also function as a transcriptional activator, depending on the stage of development and spatial restrictions. May function redundantly with odd and drm in leg joint formation during the larval stages, acting downstream of Notch activation.
Similarity
Belongs to the Odd C2H2-type zinc-finger protein family.
Keywords
Activator
Complete proteome
Developmental protein
DNA-binding
Metal-binding
Nucleus
Pair-rule protein
Reference proteome
Repeat
Repressor
Transcription
Transcription regulation
Zinc
Zinc-finger
Feature
chain Protein odd-skipped
Uniprot
H9JJ40
A0A2W1BNL9
A0A2H1X010
A0A0N1PIG0
A0A194Q0W2
A0A212ERT7
+ More
A0A0L7LCT0 A0A1Y1MTS8 A0A0T6BEZ8 A0A154PLI2 A0A1A9WHZ9 A0A0L7QNF1 A0A1A9V075 A0A1B0G5Q1 A0A1B0A815 A0A1B0AZ84 A0A1A9YR17 A0A0L0BZ98 A0A158P1R7 A0A2A3EKA1 F4WKF1 A0A195BS66 A0A088AEM3 A0A0C9RJZ8 A0A0K8TYD7 E0VDT0 A0A0K8V8I0 A0A034VMR8 A0A1L8DDA2 B4Q9M8 B4I311 B3N346 A0A1I8NQB3 P23803
A0A0L7LCT0 A0A1Y1MTS8 A0A0T6BEZ8 A0A154PLI2 A0A1A9WHZ9 A0A0L7QNF1 A0A1A9V075 A0A1B0G5Q1 A0A1B0A815 A0A1B0AZ84 A0A1A9YR17 A0A0L0BZ98 A0A158P1R7 A0A2A3EKA1 F4WKF1 A0A195BS66 A0A088AEM3 A0A0C9RJZ8 A0A0K8TYD7 E0VDT0 A0A0K8V8I0 A0A034VMR8 A0A1L8DDA2 B4Q9M8 B4I311 B3N346 A0A1I8NQB3 P23803
Pubmed
EMBL
BABH01040265
KT254332
ALR81972.1
KZ150022
PZC74877.1
ODYU01012339
+ More
SOQ58597.1 KQ460317 KPJ16018.1 KQ459583 KPI98634.1 AGBW02012932 OWR44202.1 JTDY01001744 KOB73016.1 GEZM01023093 JAV88588.1 LJIG01001026 KRT85914.1 KQ434962 KZC12693.1 KQ414864 KOC60011.1 CCAG010008430 JXJN01006207 JRES01001119 KNC25355.1 ADTU01006771 KZ288230 PBC31616.1 GL888199 EGI65375.1 KQ976419 KYM89354.1 GBYB01008525 JAG78292.1 GDHF01032815 JAI19499.1 DS235088 EEB11536.1 GDHF01017133 JAI35181.1 GAKP01015213 JAC43739.1 GFDF01009666 JAV04418.1 CM000361 CM002910 EDX03639.1 KMY87925.1 CH480820 EDW54156.1 CH954177 EDV57645.1 X57480 AE014134 BT015214
SOQ58597.1 KQ460317 KPJ16018.1 KQ459583 KPI98634.1 AGBW02012932 OWR44202.1 JTDY01001744 KOB73016.1 GEZM01023093 JAV88588.1 LJIG01001026 KRT85914.1 KQ434962 KZC12693.1 KQ414864 KOC60011.1 CCAG010008430 JXJN01006207 JRES01001119 KNC25355.1 ADTU01006771 KZ288230 PBC31616.1 GL888199 EGI65375.1 KQ976419 KYM89354.1 GBYB01008525 JAG78292.1 GDHF01032815 JAI19499.1 DS235088 EEB11536.1 GDHF01017133 JAI35181.1 GAKP01015213 JAC43739.1 GFDF01009666 JAV04418.1 CM000361 CM002910 EDX03639.1 KMY87925.1 CH480820 EDW54156.1 CH954177 EDV57645.1 X57480 AE014134 BT015214
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000037510
UP000076502
+ More
UP000091820 UP000053825 UP000078200 UP000092444 UP000092445 UP000092460 UP000092443 UP000037069 UP000005205 UP000242457 UP000007755 UP000078540 UP000005203 UP000009046 UP000000304 UP000001292 UP000008711 UP000095300 UP000000803
UP000091820 UP000053825 UP000078200 UP000092444 UP000092445 UP000092460 UP000092443 UP000037069 UP000005205 UP000242457 UP000007755 UP000078540 UP000005203 UP000009046 UP000000304 UP000001292 UP000008711 UP000095300 UP000000803
Pfam
PF00096 zf-C2H2
SUPFAM
SSF57667
SSF57667
ProteinModelPortal
H9JJ40
A0A2W1BNL9
A0A2H1X010
A0A0N1PIG0
A0A194Q0W2
A0A212ERT7
+ More
A0A0L7LCT0 A0A1Y1MTS8 A0A0T6BEZ8 A0A154PLI2 A0A1A9WHZ9 A0A0L7QNF1 A0A1A9V075 A0A1B0G5Q1 A0A1B0A815 A0A1B0AZ84 A0A1A9YR17 A0A0L0BZ98 A0A158P1R7 A0A2A3EKA1 F4WKF1 A0A195BS66 A0A088AEM3 A0A0C9RJZ8 A0A0K8TYD7 E0VDT0 A0A0K8V8I0 A0A034VMR8 A0A1L8DDA2 B4Q9M8 B4I311 B3N346 A0A1I8NQB3 P23803
A0A0L7LCT0 A0A1Y1MTS8 A0A0T6BEZ8 A0A154PLI2 A0A1A9WHZ9 A0A0L7QNF1 A0A1A9V075 A0A1B0G5Q1 A0A1B0A815 A0A1B0AZ84 A0A1A9YR17 A0A0L0BZ98 A0A158P1R7 A0A2A3EKA1 F4WKF1 A0A195BS66 A0A088AEM3 A0A0C9RJZ8 A0A0K8TYD7 E0VDT0 A0A0K8V8I0 A0A034VMR8 A0A1L8DDA2 B4Q9M8 B4I311 B3N346 A0A1I8NQB3 P23803
PDB
2EE8
E-value=3.08905e-37,
Score=388
Ontologies
GO
Topology
Subcellular location
Nucleus
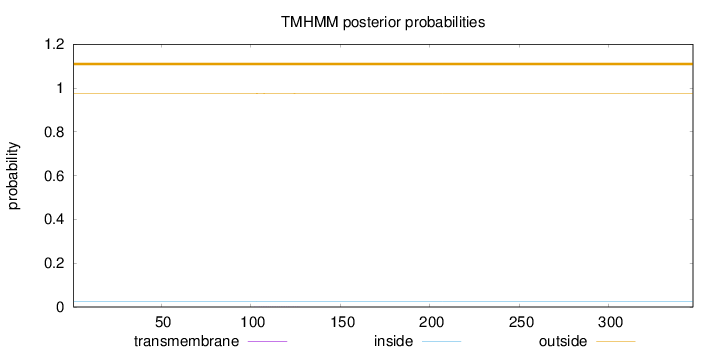
Length:
347
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01157
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02509
outside
1 - 347
Population Genetic Test Statistics
Pi
216.574328
Theta
188.018266
Tajima's D
0.558051
CLR
19.314826
CSRT
0.533123343832808
Interpretation
Uncertain
