Pre Gene Modal
BGIBMGA007493
Annotation
mitochondrial_transcription_factor_A_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 2.469
Sequence
CDS
ATGACTACTTATACTCAATTACAGCGCCTTAGTAATTATTTTTTAGGGAATTACAAGACTGTTCTATGCGGAAGAGTAAATTGGATAACGCCAATACAATCATGTGACTATACAAAAAAATCTGCAGAGCAGAGGTTGGGCCTGAACAAGCCTAAACGACCTCTTACGCCATTCTTTAAGTTCATGTCACAAATGCGACCAGCTCTTCTTGCCAAGAACCCTGGCATATCATCCAAAGAGGCTATAGCATGGACGTCGAAACATTGGCAACAACTTGATATGGAGACAAAAACCCAAATGGCGAAAGAATATCAGAAAGATTTGGAGGATTACAATAAAATTAAAGCAATGTATGAGACATCGTTAACAGAGGAACAAAAGGCAGATATAAAAAGAGTTAAAGAAGAAATGGCACAGGCCAAAGAAAAGAGAAAATTAAAAGCGGAATATAAGGAACTGGGCCGGCCTAAGAAACCAATGTCTTCATACTTTATATACATGCAGTCTAGAAAAGATAATATACAAGGAAAAACACTAAAGGAATATCAAGAGACAGTCAAGAAAGATTGGATAAACTTACCTGATAGTGAAAAAGCAAAACTGGAGAAGCAAGCACAAACATTAATGGATAAATATAAGAAAGATCTTCAAGCATGGGAACTAAAAATGGTTTCAATTGGTCGGACAGATCTAGTTAGATCGAAACCAGCCAAAGAGAAGAAAACCAAAAAAGTTGATAGTTCACAATAG
Protein
MTTYTQLQRLSNYFLGNYKTVLCGRVNWITPIQSCDYTKKSAEQRLGLNKPKRPLTPFFKFMSQMRPALLAKNPGISSKEAIAWTSKHWQQLDMETKTQMAKEYQKDLEDYNKIKAMYETSLTEEQKADIKRVKEEMAQAKEKRKLKAEYKELGRPKKPMSSYFIYMQSRKDNIQGKTLKEYQETVKKDWINLPDSEKAKLEKQAQTLMDKYKKDLQAWELKMVSIGRTDLVRSKPAKEKKTKKVDSSQ
Summary
Uniprot
Q2F674
A0A2A4JEZ8
A0A140CTX6
A0A1E1WFK5
A0A2H1WHI4
A0A2W1BMZ7
+ More
S4PDJ0 A0A194QHB3 A0A0L7KKN0 B4NFW4 A0A0Q9WUA4 A0A1W4UY31 A0A0Q9WFM2 B4M195 A0A0M4EJP4 A0A1W4VAE0 A0A1W4V9Y4 B4PPP3 Q9VDL2 B4QS21 B4IIB1 B4JXS9 B3M0C1 B4GN20 A0A0P8XZW9 A0A0R1E1A7 Q29BR2 A0A0P9ARK0 Q86BR8 A0A3B0K3Q8 B3NYX4 A0A0A1XNN0 A0A1L8ECH2 A0A1L8ECX3 A0A3B0KC55 A0A0R3NLB8 A0A0A1WHB3 B4K7R1 A0A0Q9X247 A0A067RTC2 W8AZK6 W8BYD1 A0A034VB48 A0A1I8N5Y0 A0A034VG43 A0A0K8UDH1 A0A0K8U880 A0A0K8WKX3 A0A1I8N5Z3 R4WRI6 A0A1I8PH55 C4N168 A0A1I8PGX7 A0A1J1IPZ4 A0A2J7Q5L3 A0A2J7Q5N1 A0A1B6FRB0 A0A1B6DBC7 A0A1B0DB64 D3TQN2 A0A1A9Z744 A0A0L0BUQ4 A0A1B0G2H4 A0A336M6C8 Q16FE6 A0A1S4G2W0 A0A232EH64 A0A1B0ASN8 A0A1A9WVQ1 B0W007 K7J3H3 A0A182GL54 A0A0P4VQL8 Q16IJ8 A0A2M4AYJ2 A0A023ENS6 A0A1Q3F659 A0A1Q3F5F9 A0A0V0G8I8 A0A1B6KJS9 A0A2M4AYQ6 D6WV01 W5JAX0 A0A2S2QMB2 A0A069DQW2 A0A2M3ZBB1 A0A2M3ZBF0 A0A1D2N8G9 A0A1B0CQZ6 A0A170Y569 A0A182QYD5 A0A1A9V5H5 A0A224XX71 A0A2M4BWI4 A0A1L8DHZ4 A0A166IWC7 A0A2M3ZAZ6
S4PDJ0 A0A194QHB3 A0A0L7KKN0 B4NFW4 A0A0Q9WUA4 A0A1W4UY31 A0A0Q9WFM2 B4M195 A0A0M4EJP4 A0A1W4VAE0 A0A1W4V9Y4 B4PPP3 Q9VDL2 B4QS21 B4IIB1 B4JXS9 B3M0C1 B4GN20 A0A0P8XZW9 A0A0R1E1A7 Q29BR2 A0A0P9ARK0 Q86BR8 A0A3B0K3Q8 B3NYX4 A0A0A1XNN0 A0A1L8ECH2 A0A1L8ECX3 A0A3B0KC55 A0A0R3NLB8 A0A0A1WHB3 B4K7R1 A0A0Q9X247 A0A067RTC2 W8AZK6 W8BYD1 A0A034VB48 A0A1I8N5Y0 A0A034VG43 A0A0K8UDH1 A0A0K8U880 A0A0K8WKX3 A0A1I8N5Z3 R4WRI6 A0A1I8PH55 C4N168 A0A1I8PGX7 A0A1J1IPZ4 A0A2J7Q5L3 A0A2J7Q5N1 A0A1B6FRB0 A0A1B6DBC7 A0A1B0DB64 D3TQN2 A0A1A9Z744 A0A0L0BUQ4 A0A1B0G2H4 A0A336M6C8 Q16FE6 A0A1S4G2W0 A0A232EH64 A0A1B0ASN8 A0A1A9WVQ1 B0W007 K7J3H3 A0A182GL54 A0A0P4VQL8 Q16IJ8 A0A2M4AYJ2 A0A023ENS6 A0A1Q3F659 A0A1Q3F5F9 A0A0V0G8I8 A0A1B6KJS9 A0A2M4AYQ6 D6WV01 W5JAX0 A0A2S2QMB2 A0A069DQW2 A0A2M3ZBB1 A0A2M3ZBF0 A0A1D2N8G9 A0A1B0CQZ6 A0A170Y569 A0A182QYD5 A0A1A9V5H5 A0A224XX71 A0A2M4BWI4 A0A1L8DHZ4 A0A166IWC7 A0A2M3ZAZ6
Pubmed
28756777
23622113
26354079
26227816
17994087
17550304
+ More
10731132 11554753 11171100 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 25830018 24845553 24495485 25348373 25315136 23691247 19576987 20353571 26108605 17510324 28648823 20075255 26483478 27129103 24945155 18362917 19820115 20920257 23761445 26334808 27289101
10731132 11554753 11171100 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 25830018 24845553 24495485 25348373 25315136 23691247 19576987 20353571 26108605 17510324 28648823 20075255 26483478 27129103 24945155 18362917 19820115 20920257 23761445 26334808 27289101
EMBL
DQ311198
ABD36143.2
NWSH01001823
PCG70003.1
KU552107
AMH87783.1
+ More
GDQN01005316 JAT85738.1 ODYU01008711 SOQ52529.1 KZ149958 PZC76429.1 GAIX01004932 JAA87628.1 KQ458860 KPJ04809.1 JTDY01009331 KOB63645.1 CH964251 EDW83181.1 KRF99743.1 CH940650 KRF83320.1 EDW67506.1 CP012526 ALC47196.1 CM000160 EDW96143.1 AY061457 AE014297 AB045318 AB040745 AAL29005.1 AAN14356.1 BAB20089.1 BAB62223.1 CM000364 EDX12217.1 CH480842 EDW49655.1 CH916377 EDV90491.1 CH902617 EDV43127.1 CH479186 EDW39146.1 KPU80095.1 KRK02937.1 CM000070 EAL26935.2 KPU80094.1 AAF55779.2 OUUW01000007 SPP82580.1 CH954181 EDV48377.1 GBXI01001368 JAD12924.1 GFDG01002369 JAV16430.1 GFDG01002370 JAV16429.1 SPP82581.1 KRS99669.1 GBXI01016519 JAC97772.1 CH933806 EDW16432.1 KRG02124.1 KK852428 KDR24060.1 GAMC01012287 GAMC01012285 JAB94270.1 GAMC01012286 JAB94269.1 GAKP01018416 JAC40536.1 GAKP01018414 JAC40538.1 GDHF01027602 JAI24712.1 GDHF01029531 GDHF01028329 JAI22783.1 JAI23985.1 GDHF01013098 GDHF01000610 JAI39216.1 JAI51704.1 AK417302 BAN20517.1 EZ048841 ACN69133.1 CVRI01000056 CRL01794.1 NEVH01017557 PNF23876.1 PNF23875.1 GECZ01017022 JAS52747.1 GEDC01018233 GEDC01014393 JAS19065.1 JAS22905.1 AJVK01004632 EZ423734 ADD20010.1 JRES01001300 KNC23800.1 CCAG010018314 UFQS01000618 UFQT01000618 SSX05457.1 SSX25816.1 CH478458 EAT32963.1 EAT32964.1 NNAY01004596 OXU17674.1 JXJN01002890 DS231816 EDS37579.1 JXUM01071245 KQ562651 KXJ75407.1 GDKW01002457 JAI54138.1 CH478073 EAT34084.1 GGFK01012520 MBW45841.1 GAPW01003504 JAC10094.1 GFDL01011990 JAV23055.1 GFDL01012248 JAV22797.1 GECL01001673 JAP04451.1 GEBQ01028255 GEBQ01022696 JAT11722.1 JAT17281.1 GGFK01012521 MBW45842.1 KQ971357 EFA08515.1 ADMH02001646 ETN61607.1 GGMS01009661 MBY78864.1 GBGD01002793 JAC86096.1 GGFM01005030 MBW25781.1 GGFM01005115 MBW25866.1 LJIJ01000147 ODN01559.1 AJWK01024249 GEMB01003654 JAR99564.1 AXCN02000647 GFTR01003835 JAW12591.1 GGFJ01008221 MBW57362.1 GFDF01008008 JAV06076.1 KU364383 ANA11557.1 GGFM01004932 MBW25683.1
GDQN01005316 JAT85738.1 ODYU01008711 SOQ52529.1 KZ149958 PZC76429.1 GAIX01004932 JAA87628.1 KQ458860 KPJ04809.1 JTDY01009331 KOB63645.1 CH964251 EDW83181.1 KRF99743.1 CH940650 KRF83320.1 EDW67506.1 CP012526 ALC47196.1 CM000160 EDW96143.1 AY061457 AE014297 AB045318 AB040745 AAL29005.1 AAN14356.1 BAB20089.1 BAB62223.1 CM000364 EDX12217.1 CH480842 EDW49655.1 CH916377 EDV90491.1 CH902617 EDV43127.1 CH479186 EDW39146.1 KPU80095.1 KRK02937.1 CM000070 EAL26935.2 KPU80094.1 AAF55779.2 OUUW01000007 SPP82580.1 CH954181 EDV48377.1 GBXI01001368 JAD12924.1 GFDG01002369 JAV16430.1 GFDG01002370 JAV16429.1 SPP82581.1 KRS99669.1 GBXI01016519 JAC97772.1 CH933806 EDW16432.1 KRG02124.1 KK852428 KDR24060.1 GAMC01012287 GAMC01012285 JAB94270.1 GAMC01012286 JAB94269.1 GAKP01018416 JAC40536.1 GAKP01018414 JAC40538.1 GDHF01027602 JAI24712.1 GDHF01029531 GDHF01028329 JAI22783.1 JAI23985.1 GDHF01013098 GDHF01000610 JAI39216.1 JAI51704.1 AK417302 BAN20517.1 EZ048841 ACN69133.1 CVRI01000056 CRL01794.1 NEVH01017557 PNF23876.1 PNF23875.1 GECZ01017022 JAS52747.1 GEDC01018233 GEDC01014393 JAS19065.1 JAS22905.1 AJVK01004632 EZ423734 ADD20010.1 JRES01001300 KNC23800.1 CCAG010018314 UFQS01000618 UFQT01000618 SSX05457.1 SSX25816.1 CH478458 EAT32963.1 EAT32964.1 NNAY01004596 OXU17674.1 JXJN01002890 DS231816 EDS37579.1 JXUM01071245 KQ562651 KXJ75407.1 GDKW01002457 JAI54138.1 CH478073 EAT34084.1 GGFK01012520 MBW45841.1 GAPW01003504 JAC10094.1 GFDL01011990 JAV23055.1 GFDL01012248 JAV22797.1 GECL01001673 JAP04451.1 GEBQ01028255 GEBQ01022696 JAT11722.1 JAT17281.1 GGFK01012521 MBW45842.1 KQ971357 EFA08515.1 ADMH02001646 ETN61607.1 GGMS01009661 MBY78864.1 GBGD01002793 JAC86096.1 GGFM01005030 MBW25781.1 GGFM01005115 MBW25866.1 LJIJ01000147 ODN01559.1 AJWK01024249 GEMB01003654 JAR99564.1 AXCN02000647 GFTR01003835 JAW12591.1 GGFJ01008221 MBW57362.1 GFDF01008008 JAV06076.1 KU364383 ANA11557.1 GGFM01004932 MBW25683.1
Proteomes
UP000218220
UP000053268
UP000037510
UP000007798
UP000192221
UP000008792
+ More
UP000092553 UP000002282 UP000000803 UP000000304 UP000001292 UP000001070 UP000007801 UP000008744 UP000001819 UP000268350 UP000008711 UP000009192 UP000027135 UP000095301 UP000095300 UP000183832 UP000235965 UP000092462 UP000092445 UP000037069 UP000092444 UP000008820 UP000215335 UP000092460 UP000091820 UP000002320 UP000002358 UP000069940 UP000249989 UP000007266 UP000000673 UP000094527 UP000092461 UP000075886 UP000078200
UP000092553 UP000002282 UP000000803 UP000000304 UP000001292 UP000001070 UP000007801 UP000008744 UP000001819 UP000268350 UP000008711 UP000009192 UP000027135 UP000095301 UP000095300 UP000183832 UP000235965 UP000092462 UP000092445 UP000037069 UP000092444 UP000008820 UP000215335 UP000092460 UP000091820 UP000002320 UP000002358 UP000069940 UP000249989 UP000007266 UP000000673 UP000094527 UP000092461 UP000075886 UP000078200
SUPFAM
SSF47095
SSF47095
Gene 3D
ProteinModelPortal
Q2F674
A0A2A4JEZ8
A0A140CTX6
A0A1E1WFK5
A0A2H1WHI4
A0A2W1BMZ7
+ More
S4PDJ0 A0A194QHB3 A0A0L7KKN0 B4NFW4 A0A0Q9WUA4 A0A1W4UY31 A0A0Q9WFM2 B4M195 A0A0M4EJP4 A0A1W4VAE0 A0A1W4V9Y4 B4PPP3 Q9VDL2 B4QS21 B4IIB1 B4JXS9 B3M0C1 B4GN20 A0A0P8XZW9 A0A0R1E1A7 Q29BR2 A0A0P9ARK0 Q86BR8 A0A3B0K3Q8 B3NYX4 A0A0A1XNN0 A0A1L8ECH2 A0A1L8ECX3 A0A3B0KC55 A0A0R3NLB8 A0A0A1WHB3 B4K7R1 A0A0Q9X247 A0A067RTC2 W8AZK6 W8BYD1 A0A034VB48 A0A1I8N5Y0 A0A034VG43 A0A0K8UDH1 A0A0K8U880 A0A0K8WKX3 A0A1I8N5Z3 R4WRI6 A0A1I8PH55 C4N168 A0A1I8PGX7 A0A1J1IPZ4 A0A2J7Q5L3 A0A2J7Q5N1 A0A1B6FRB0 A0A1B6DBC7 A0A1B0DB64 D3TQN2 A0A1A9Z744 A0A0L0BUQ4 A0A1B0G2H4 A0A336M6C8 Q16FE6 A0A1S4G2W0 A0A232EH64 A0A1B0ASN8 A0A1A9WVQ1 B0W007 K7J3H3 A0A182GL54 A0A0P4VQL8 Q16IJ8 A0A2M4AYJ2 A0A023ENS6 A0A1Q3F659 A0A1Q3F5F9 A0A0V0G8I8 A0A1B6KJS9 A0A2M4AYQ6 D6WV01 W5JAX0 A0A2S2QMB2 A0A069DQW2 A0A2M3ZBB1 A0A2M3ZBF0 A0A1D2N8G9 A0A1B0CQZ6 A0A170Y569 A0A182QYD5 A0A1A9V5H5 A0A224XX71 A0A2M4BWI4 A0A1L8DHZ4 A0A166IWC7 A0A2M3ZAZ6
S4PDJ0 A0A194QHB3 A0A0L7KKN0 B4NFW4 A0A0Q9WUA4 A0A1W4UY31 A0A0Q9WFM2 B4M195 A0A0M4EJP4 A0A1W4VAE0 A0A1W4V9Y4 B4PPP3 Q9VDL2 B4QS21 B4IIB1 B4JXS9 B3M0C1 B4GN20 A0A0P8XZW9 A0A0R1E1A7 Q29BR2 A0A0P9ARK0 Q86BR8 A0A3B0K3Q8 B3NYX4 A0A0A1XNN0 A0A1L8ECH2 A0A1L8ECX3 A0A3B0KC55 A0A0R3NLB8 A0A0A1WHB3 B4K7R1 A0A0Q9X247 A0A067RTC2 W8AZK6 W8BYD1 A0A034VB48 A0A1I8N5Y0 A0A034VG43 A0A0K8UDH1 A0A0K8U880 A0A0K8WKX3 A0A1I8N5Z3 R4WRI6 A0A1I8PH55 C4N168 A0A1I8PGX7 A0A1J1IPZ4 A0A2J7Q5L3 A0A2J7Q5N1 A0A1B6FRB0 A0A1B6DBC7 A0A1B0DB64 D3TQN2 A0A1A9Z744 A0A0L0BUQ4 A0A1B0G2H4 A0A336M6C8 Q16FE6 A0A1S4G2W0 A0A232EH64 A0A1B0ASN8 A0A1A9WVQ1 B0W007 K7J3H3 A0A182GL54 A0A0P4VQL8 Q16IJ8 A0A2M4AYJ2 A0A023ENS6 A0A1Q3F659 A0A1Q3F5F9 A0A0V0G8I8 A0A1B6KJS9 A0A2M4AYQ6 D6WV01 W5JAX0 A0A2S2QMB2 A0A069DQW2 A0A2M3ZBB1 A0A2M3ZBF0 A0A1D2N8G9 A0A1B0CQZ6 A0A170Y569 A0A182QYD5 A0A1A9V5H5 A0A224XX71 A0A2M4BWI4 A0A1L8DHZ4 A0A166IWC7 A0A2M3ZAZ6
PDB
3TMM
E-value=1.5095e-16,
Score=208
Ontologies
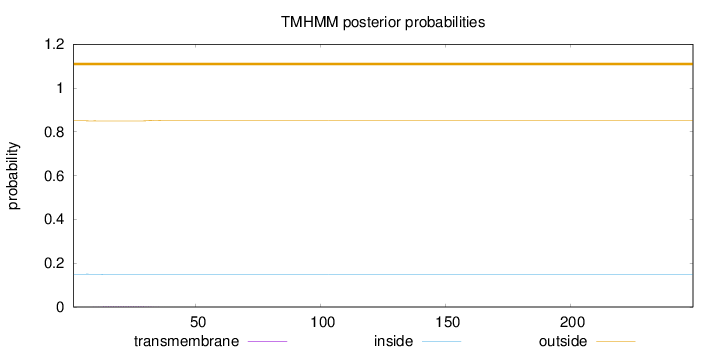
Topology
Length:
249
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06341
Exp number, first 60 AAs:
0.06324
Total prob of N-in:
0.14995
outside
1 - 249
Population Genetic Test Statistics
Pi
167.655846
Theta
160.795926
Tajima's D
0.214617
CLR
0.800768
CSRT
0.427978601069947
Interpretation
Uncertain
