Gene
KWMTBOMO00902
Pre Gene Modal
BGIBMGA009653
Annotation
PREDICTED:_transcription_factor_CP2-like_protein_1_isoform_X1_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 4.091
Sequence
CDS
ATGGAATATACGGAATATAGAGACTTTACAATGCATGACGTACGTAAAGAGGAGTACGGCTGGGAATCTACAGTTAGACTCATGGTCGGGAGGGAGGACGAATTGGGCATGTACCTGGAGGAGAGCAGGGAGGGCAGGAAGCGGAAGTGGCAGCCCGAGAAAATGAGGCGAGACGGCAGCGAAGATAAAGATCAGAAGAAGGTGCCTCCGAAGAAGATCATGGCTGCCCTAAGCGAGATAGGCGGGAGCGGCACAAGCTCCTCATCCTCGCATTTGTCTCCGGCATGGCAAGTCAACGAACTCGATCTGGACTTGCCCGTCGAACTATCAATGAACGAAGCGCTGTTGTCGCTTCCATCGTTGGCGGTGTTCAAACAGGAGGCTCCGTCCCCGACGGGCGCGGTGCTCTCGCCCCCCCGCCGCCACTGGCCCGTCCCCCGACGGAACGACGATCGGCAGATGACAAGCATGGTTGTCGACGGAGCCCGTGACGGCATGGACGAGTGCCAGCAGTCGCCTTCCCACAACGGAAACGCTATGCAGCATGGAAGTCCCGACGCCTTGCAGTGCCAACAGCAGATGCCGCCGAGCGCCGTCATGCCAATGAACGGTTACCATTCACCAAACGAACACGAGAATAAAAACGCTGGTCTGCTGATCTGCTCCCCGGTGGGCTCGCTGGAGGGCTACCTGCACTCTCCACCACACCTGCAGCCGCGCTCCGACTCCAGCTTCAAGGATGACACCAGGTTCCAGTACGTTCTGGCCGCAGCGACTTCGATAGCAACCAAACAGAACGAGGAAACCCTGACCTACCTCAACCAAGGCCAGTCCTACGAGATCAAGCTCAAGAAGCTCGGAGACTTGTCACATTACAAAGGAAAGCTGCTGAAAAGTGTCATCAAGATATGCTTCCACGAACGCCGCTTGCAGTACATGGAAAGGGAGCAGATAGCGCAGTGGCACGCCGAGAGACCGGGGGAGAGGATCGTAGAAGTGGACGTGCCTCTGTCGTACGGAATTATAAAAGTGGAACAATCCACAGCACTGAACGCGCTCCACGTCTACTGGGACGCCACCAAGGAAGTGGGTGTTTATATAAAGGTGAACTGCATCTCAACGGAATTCACAGCCAAGAAGCACGGCGGCGAGAAGGGAGTCCCATTCCGTATACAGGTCGAGACTTATCTCGCGAGCGACGAGAACAATCGTCTGAACGCCGCCGCCTGCCAGATCAAGGTATTCAAATTGAAAGGCGCCGATCGCAAACACAAACAGGACAGGGAGAAGGTGCTGCGCAGACCGAGGGCCGATCTGGACAAGTACCAGCCCGCCTGCGAGGCCACTGTGCTGACCACGTTGTCACACGACGCGCTGATGCCGCCGCCCTCGCTGGTCACCACCTCGCCGTCCTACTCGCCAGAGCTGTGTGTGACACCTAAATCTGCCTCGAGCCCTATCACTACCAAGCCGATTTCTCAACCAGCCATCGTTGGTACTACTAATCAAATGAAGCCGCTGTATTGTCAAGATGCTGGTGAGATGAAGGTGAGCAGCCCAACCTGCCACTCCCCAGGAGCTATCCTGCCTGATCTACCACAGCCTGCTGAGGAACCAGACAAGAACTTTGGATTATCAAAAGATGCCACCTCCGCCGAGACTCAGGTGTGGTTGTCCCGACAACGGTTCATGCAACACGCCGCCACGTTCGCGAACTTTTCCGGGGCTGATCTGCTGAGACTATCCCGTGATGACATTATCCAGATCGTCGGACTGGCTGATGGCATACGTCTGTTCAACGCACTTCACGCCAAACGCATCTCGCCGCGCCTCACCCTGTACGTGTGCCCGCCCTCCTCCGACGTGCACTGCGCGCTCTACCTGCACGCGTGTCGCGCGCACGAGCTGGTGCACAAGCTGAACGCGCTGCAGGGAGCCGGCAGCAAGGATTGCGAAACCGTATTGGTGTCCGGTCCGAACGGCGCTCGCGTCCGCCTCACCGACGAGCTGGTGCGACATCTGCCAGACAACTCGACTTATCGCCTCAAGAGACTCGAAGACTGCAATGCTCTGCTCCTAAGCGCCACCACCGGCAACTGA
Protein
MEYTEYRDFTMHDVRKEEYGWESTVRLMVGREDELGMYLEESREGRKRKWQPEKMRRDGSEDKDQKKVPPKKIMAALSEIGGSGTSSSSSHLSPAWQVNELDLDLPVELSMNEALLSLPSLAVFKQEAPSPTGAVLSPPRRHWPVPRRNDDRQMTSMVVDGARDGMDECQQSPSHNGNAMQHGSPDALQCQQQMPPSAVMPMNGYHSPNEHENKNAGLLICSPVGSLEGYLHSPPHLQPRSDSSFKDDTRFQYVLAAATSIATKQNEETLTYLNQGQSYEIKLKKLGDLSHYKGKLLKSVIKICFHERRLQYMEREQIAQWHAERPGERIVEVDVPLSYGIIKVEQSTALNALHVYWDATKEVGVYIKVNCISTEFTAKKHGGEKGVPFRIQVETYLASDENNRLNAAACQIKVFKLKGADRKHKQDREKVLRRPRADLDKYQPACEATVLTTLSHDALMPPPSLVTTSPSYSPELCVTPKSASSPITTKPISQPAIVGTTNQMKPLYCQDAGEMKVSSPTCHSPGAILPDLPQPAEEPDKNFGLSKDATSAETQVWLSRQRFMQHAATFANFSGADLLRLSRDDIIQIVGLADGIRLFNALHAKRISPRLTLYVCPPSSDVHCALYLHACRAHELVHKLNALQGAGSKDCETVLVSGPNGARVRLTDELVRHLPDNSTYRLKRLEDCNALLLSATTGN
Summary
Uniprot
A0A2J7Q2S2
A0A2J7Q2R8
A0A139WF83
A0A2J7Q2S9
A0A1Y1LDP9
A0A1Y1LA11
+ More
V5G4Q8 A0A1L8DIC0 A0A1L8DIH0 A0A1L8DI50 A0A1L8DI60 A0A1B6DG69 A0A1B6GEK0 A0A023EVF9 A0A0J7L5F1 Q17K77 A0A1B0CIF9 A0A232FAA7 A0A158N974 A0A1S4G817 A0A0P5BDU5 A0A0V0G599 A0A224X9T8 L7ML72 A0A1J1IB54 A0A0A9Z1B5 A0A182JRP9 A0A0A9Z2U4 A0A182RMZ7 A0A293MTN9 A0A182WL00 B4J6U9 B4QI95
V5G4Q8 A0A1L8DIC0 A0A1L8DIH0 A0A1L8DI50 A0A1L8DI60 A0A1B6DG69 A0A1B6GEK0 A0A023EVF9 A0A0J7L5F1 Q17K77 A0A1B0CIF9 A0A232FAA7 A0A158N974 A0A1S4G817 A0A0P5BDU5 A0A0V0G599 A0A224X9T8 L7ML72 A0A1J1IB54 A0A0A9Z1B5 A0A182JRP9 A0A0A9Z2U4 A0A182RMZ7 A0A293MTN9 A0A182WL00 B4J6U9 B4QI95
Pubmed
EMBL
NEVH01019077
PNF22878.1
PNF22880.1
KQ971354
KYB26457.1
PNF22879.1
+ More
GEZM01062471 GEZM01062470 JAV69706.1 GEZM01062469 JAV69708.1 GALX01003456 JAB65010.1 GFDF01007959 JAV06125.1 GFDF01007960 JAV06124.1 GFDF01007958 JAV06126.1 GFDF01007961 JAV06123.1 GEDC01014135 GEDC01012621 GEDC01010142 JAS23163.1 JAS24677.1 JAS27156.1 GECZ01013467 GECZ01008897 GECZ01006525 GECZ01005691 JAS56302.1 JAS60872.1 JAS63244.1 JAS64078.1 GAPW01000612 JAC12986.1 LBMM01000662 KMQ97851.1 CH477227 EAT47108.1 AJWK01013295 AJWK01013296 AJWK01013297 AJWK01013298 AJWK01013299 AJWK01013300 AJWK01013301 AJWK01013302 AJWK01013303 AJWK01013304 NNAY01000542 OXU27781.1 ADTU01008785 ADTU01008786 ADTU01008787 ADTU01008788 ADTU01008789 GDIP01191218 JAJ32184.1 GECL01002910 JAP03214.1 GFTR01007341 JAW09085.1 GACK01001110 JAA63924.1 CVRI01000042 CRK95673.1 GBHO01035091 GBHO01005953 GBHO01005947 GBRD01007938 GBRD01007937 JAG08513.1 JAG37651.1 JAG37657.1 JAG57883.1 GBHO01005951 GBHO01005949 GBHO01005948 GBRD01007941 GBRD01007940 JAG37653.1 JAG37655.1 JAG37656.1 JAG57880.1 GFWV01018613 MAA43341.1 CH916367 EDW02030.1 CM000362 EDX06455.1
GEZM01062471 GEZM01062470 JAV69706.1 GEZM01062469 JAV69708.1 GALX01003456 JAB65010.1 GFDF01007959 JAV06125.1 GFDF01007960 JAV06124.1 GFDF01007958 JAV06126.1 GFDF01007961 JAV06123.1 GEDC01014135 GEDC01012621 GEDC01010142 JAS23163.1 JAS24677.1 JAS27156.1 GECZ01013467 GECZ01008897 GECZ01006525 GECZ01005691 JAS56302.1 JAS60872.1 JAS63244.1 JAS64078.1 GAPW01000612 JAC12986.1 LBMM01000662 KMQ97851.1 CH477227 EAT47108.1 AJWK01013295 AJWK01013296 AJWK01013297 AJWK01013298 AJWK01013299 AJWK01013300 AJWK01013301 AJWK01013302 AJWK01013303 AJWK01013304 NNAY01000542 OXU27781.1 ADTU01008785 ADTU01008786 ADTU01008787 ADTU01008788 ADTU01008789 GDIP01191218 JAJ32184.1 GECL01002910 JAP03214.1 GFTR01007341 JAW09085.1 GACK01001110 JAA63924.1 CVRI01000042 CRK95673.1 GBHO01035091 GBHO01005953 GBHO01005947 GBRD01007938 GBRD01007937 JAG08513.1 JAG37651.1 JAG37657.1 JAG57883.1 GBHO01005951 GBHO01005949 GBHO01005948 GBRD01007941 GBRD01007940 JAG37653.1 JAG37655.1 JAG37656.1 JAG57880.1 GFWV01018613 MAA43341.1 CH916367 EDW02030.1 CM000362 EDX06455.1
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2J7Q2S2
A0A2J7Q2R8
A0A139WF83
A0A2J7Q2S9
A0A1Y1LDP9
A0A1Y1LA11
+ More
V5G4Q8 A0A1L8DIC0 A0A1L8DIH0 A0A1L8DI50 A0A1L8DI60 A0A1B6DG69 A0A1B6GEK0 A0A023EVF9 A0A0J7L5F1 Q17K77 A0A1B0CIF9 A0A232FAA7 A0A158N974 A0A1S4G817 A0A0P5BDU5 A0A0V0G599 A0A224X9T8 L7ML72 A0A1J1IB54 A0A0A9Z1B5 A0A182JRP9 A0A0A9Z2U4 A0A182RMZ7 A0A293MTN9 A0A182WL00 B4J6U9 B4QI95
V5G4Q8 A0A1L8DIC0 A0A1L8DIH0 A0A1L8DI50 A0A1L8DI60 A0A1B6DG69 A0A1B6GEK0 A0A023EVF9 A0A0J7L5F1 Q17K77 A0A1B0CIF9 A0A232FAA7 A0A158N974 A0A1S4G817 A0A0P5BDU5 A0A0V0G599 A0A224X9T8 L7ML72 A0A1J1IB54 A0A0A9Z1B5 A0A182JRP9 A0A0A9Z2U4 A0A182RMZ7 A0A293MTN9 A0A182WL00 B4J6U9 B4QI95
PDB
5MR7
E-value=2.32279e-25,
Score=289
Ontologies
PANTHER
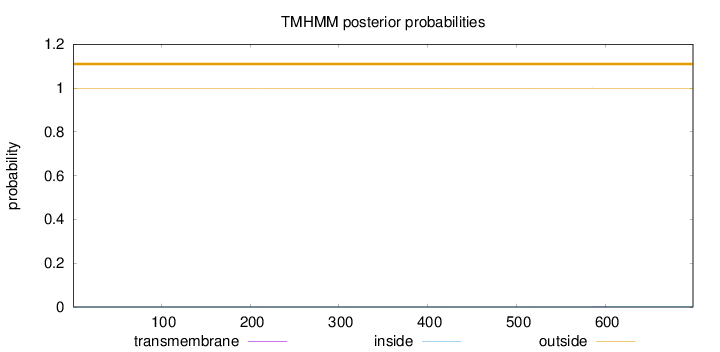
Topology
Length:
699
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00471
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00051
outside
1 - 699
Population Genetic Test Statistics
Pi
241.362727
Theta
160.468581
Tajima's D
1.718896
CLR
0.01763
CSRT
0.827108644567772
Interpretation
Uncertain
