Gene
KWMTBOMO00707
Pre Gene Modal
BGIBMGA003226
Annotation
PREDICTED:_gastrula_zinc_finger_protein_XlCGF26.1-like_[Plutella_xylostella]
Transcription factor
Location in the cell
Nuclear Reliability : 4.623
Sequence
CDS
ATGACCGAAGAGAAAAGCTATTGCAGGCTTTGCGCTGAAGCGACTTCGAACAATCAGCTTTTAGCCGCTGAAGATGTCGACGGAATAAATTTTAAAATATTAAAGAAAATGCAGTGGATAAACGTTGATGTTTCTAACGATTTACTACCGAACGCTATTTGTTTCGCGTGTTTCGACTCCTTAGAACGTACGTGGATATTTTTGCACAACGTCAGAGCGGCTCAAAAGAAATTGGCAAAGATCTTTTCTTGCGATAACAATGTGCATACAACGAATGAGGACGTTGGTCGACCTGTCGATTGCGACTGGGAGGTTCTGACCGCGGCCAACGTAAAAGAAGAATCCGAGGTGCCCACAAATAATTTCGAAAATGAAAGAGCCGCTGAAAAGAAATTAGCACATATCCTTTCTTGCGATAACAGAGTGCATACACCTAATGAGGACGCTGGTCGTCCTATCGATTGCGAGTGGGAGGTCCTGACCGCGGCTAATGTAAAAGAAGAATCCGACGAGCCCACAAATAATTTTGAAGATGAAATGCTCATAGAAATCAATGAAATAAAAATCGAGAAGACAAGTGACATTGAAACTGACAGCGACATACCATTGCTACAAACAAGGGTGAAAAAGAAAAAAAAGAAAAAAAAACTAAAAAACATTGTACACATTACATCGGAAGATATAACCTGGGATGATTACACGAGCCTCTGTGCAAACTGTGACGCCCAATGCATGACCATAGCAGCCCTACGATCACATTCTTTAGAGAAACACGCTCGTTGCTTTGTATCAAAATGTCTGATCTGTGCAAAAATATCTTCTAATTATCGTTTATTAATAAACCATGCCCGAAGTCACAATCAAATGTTAAATTACCACTGTGAATACTGCAATAAATATATACCGTCAATCAGTGAACTAAAGAAACATAGGAACACGGCACACCAACATGTTTACAAGACTTACTGTCACAATTGTGGTGAGAAATTTGATACGCCGGAATCTTTAGAAGATCACATGATCGCATATTCAAACTCTTACAAAAAATGCGGTAGGAGATCGACAAAAATTAGGCCTCACAATCTTAAATGTGATATGTGCAGCAGAGACTTCAAGTCATTAAGTAATCTAATTCAACACAAAAAGGTCCACATGGAACGAACCAGAGACTTTGGCTGTCACAAATGTGGTAAAATGTTCTTTACGAAGGGAGCCCTGGTGAACCATTTGGCGATTCACGAGGAAGTGAGACCGCACAAATGTGAGTACTGCCCTCTGGCCTTCCGGGCTCGGGGGAACCTGCAGGCCCACCTCGTGTTGCACTCAGGATCGAAGCCGTTTGTGTGCGAACAGTGCGGGAAGAGTTTCCGTATGAAGAGACACTTGAAATCTCATTCCATTATGCATACAGATTTGATGCCGTATGCATGCGAGTTCTGCAATAAATTGTTTAGGTTTAAAACAAGATTGAATCTGCATTTGAGACAGCACACCGGAGCCAAGCCCTACAATTGCATTCAGTGTCAAAGAGACTTCACGAACGGTTCAAATTTCAAAAAACATATGAAACGTAGACACAACATTGATATATCAAAGAAGAAAAAATATACAATAGTAGATAAACCTGATGAGAAAATCGAAAGTGAAGAAGGAAAGGCTTGA
Protein
MTEEKSYCRLCAEATSNNQLLAAEDVDGINFKILKKMQWINVDVSNDLLPNAICFACFDSLERTWIFLHNVRAAQKKLAKIFSCDNNVHTTNEDVGRPVDCDWEVLTAANVKEESEVPTNNFENERAAEKKLAHILSCDNRVHTPNEDAGRPIDCEWEVLTAANVKEESDEPTNNFEDEMLIEINEIKIEKTSDIETDSDIPLLQTRVKKKKKKKKLKNIVHITSEDITWDDYTSLCANCDAQCMTIAALRSHSLEKHARCFVSKCLICAKISSNYRLLINHARSHNQMLNYHCEYCNKYIPSISELKKHRNTAHQHVYKTYCHNCGEKFDTPESLEDHMIAYSNSYKKCGRRSTKIRPHNLKCDMCSRDFKSLSNLIQHKKVHMERTRDFGCHKCGKMFFTKGALVNHLAIHEEVRPHKCEYCPLAFRARGNLQAHLVLHSGSKPFVCEQCGKSFRMKRHLKSHSIMHTDLMPYACEFCNKLFRFKTRLNLHLRQHTGAKPYNCIQCQRDFTNGSNFKKHMKRRHNIDISKKKKYTIVDKPDEKIESEEGKA
Summary
Uniprot
H9J139
A0A2H1VE60
A0A2A4IW19
A0A0N1IEG0
A0A0L7LDT9
A0A212FAD0
+ More
A0A194PUN4 H9J196 A0A0N1IFZ7 A0A194PVA9 A0A2A4IYH7 A0A2H1VDP3 A0A0N0PBG2 A0A2H1VDI8 D6WM19 A0A2H1VF55 A0A0C9R581 A0A0J7N9Q1 A0A1B6FUI1 A0A158NLD7 A0A310SQL1 A0A2A3E268 K7JT31 A0A195B2A8 A0A0A9WIK4 A0A195CZP3 A0A0A9WB99 A0A0V0GCQ2 A0A232FB98 A0A0L7LHP3 A0A069DVT3 N6U6U8 A0A2A4JCH3 A0A0M9A4M1 A0A0L0CPS4 A0A0K8VAN7 A0A0K8V2C5 A0A0A1XDV5 A0A147AJZ0 A0A146Q6G1 A0A147AFX3 A0A146SBW7 A0A146RL18 A0A146Q4P7
A0A194PUN4 H9J196 A0A0N1IFZ7 A0A194PVA9 A0A2A4IYH7 A0A2H1VDP3 A0A0N0PBG2 A0A2H1VDI8 D6WM19 A0A2H1VF55 A0A0C9R581 A0A0J7N9Q1 A0A1B6FUI1 A0A158NLD7 A0A310SQL1 A0A2A3E268 K7JT31 A0A195B2A8 A0A0A9WIK4 A0A195CZP3 A0A0A9WB99 A0A0V0GCQ2 A0A232FB98 A0A0L7LHP3 A0A069DVT3 N6U6U8 A0A2A4JCH3 A0A0M9A4M1 A0A0L0CPS4 A0A0K8VAN7 A0A0K8V2C5 A0A0A1XDV5 A0A147AJZ0 A0A146Q6G1 A0A147AFX3 A0A146SBW7 A0A146RL18 A0A146Q4P7
Pubmed
EMBL
BABH01031441
ODYU01001823
SOQ38554.1
NWSH01005726
PCG64005.1
KQ460940
+ More
KPJ10633.1 JTDY01001547 KOB73570.1 AGBW02009496 OWR50687.1 KQ459593 KPI96693.1 BABH01031482 KPJ10603.1 KPI96694.1 NWSH01005474 PCG64173.1 ODYU01001969 SOQ38896.1 KPJ10604.1 SOQ38895.1 KQ971343 EFA04207.1 SOQ38894.1 GBYB01002000 GBYB01002001 JAG71767.1 JAG71768.1 LBMM01007818 KMQ89400.1 GECZ01015909 JAS53860.1 ADTU01019320 KQ760118 OAD61926.1 KZ288430 PBC25833.1 KQ976662 KYM78427.1 GBHO01038929 GBHO01025727 GBHO01019226 GBHO01011035 GDHC01017998 JAG04675.1 JAG17877.1 JAG24378.1 JAG32569.1 JAQ00631.1 KQ977115 KYN05609.1 GBHO01038932 GBHO01038928 GBHO01038927 GBHO01025725 GDHC01010384 JAG04672.1 JAG04676.1 JAG04677.1 JAG17879.1 JAQ08245.1 GECL01000480 JAP05644.1 NNAY01000478 OXU28111.1 JTDY01001068 KOB74972.1 GBGD01001082 JAC87807.1 APGK01037606 KB740948 ENN77375.1 NWSH01002022 PCG69398.1 KQ435757 KOX75843.1 JRES01000091 KNC34187.1 GDHF01016391 JAI35923.1 GDHF01019361 JAI32953.1 GBXI01015391 GBXI01005162 JAC98900.1 JAD09130.1 GCES01007572 JAR78751.1 GCES01134453 JAQ51869.1 GCES01008853 JAR77470.1 GCES01108251 JAQ78071.1 GCES01117194 JAQ69128.1 GCES01135348 JAQ50974.1
KPJ10633.1 JTDY01001547 KOB73570.1 AGBW02009496 OWR50687.1 KQ459593 KPI96693.1 BABH01031482 KPJ10603.1 KPI96694.1 NWSH01005474 PCG64173.1 ODYU01001969 SOQ38896.1 KPJ10604.1 SOQ38895.1 KQ971343 EFA04207.1 SOQ38894.1 GBYB01002000 GBYB01002001 JAG71767.1 JAG71768.1 LBMM01007818 KMQ89400.1 GECZ01015909 JAS53860.1 ADTU01019320 KQ760118 OAD61926.1 KZ288430 PBC25833.1 KQ976662 KYM78427.1 GBHO01038929 GBHO01025727 GBHO01019226 GBHO01011035 GDHC01017998 JAG04675.1 JAG17877.1 JAG24378.1 JAG32569.1 JAQ00631.1 KQ977115 KYN05609.1 GBHO01038932 GBHO01038928 GBHO01038927 GBHO01025725 GDHC01010384 JAG04672.1 JAG04676.1 JAG04677.1 JAG17879.1 JAQ08245.1 GECL01000480 JAP05644.1 NNAY01000478 OXU28111.1 JTDY01001068 KOB74972.1 GBGD01001082 JAC87807.1 APGK01037606 KB740948 ENN77375.1 NWSH01002022 PCG69398.1 KQ435757 KOX75843.1 JRES01000091 KNC34187.1 GDHF01016391 JAI35923.1 GDHF01019361 JAI32953.1 GBXI01015391 GBXI01005162 JAC98900.1 JAD09130.1 GCES01007572 JAR78751.1 GCES01134453 JAQ51869.1 GCES01008853 JAR77470.1 GCES01108251 JAQ78071.1 GCES01117194 JAQ69128.1 GCES01135348 JAQ50974.1
Proteomes
SUPFAM
SSF57667
SSF57667
ProteinModelPortal
H9J139
A0A2H1VE60
A0A2A4IW19
A0A0N1IEG0
A0A0L7LDT9
A0A212FAD0
+ More
A0A194PUN4 H9J196 A0A0N1IFZ7 A0A194PVA9 A0A2A4IYH7 A0A2H1VDP3 A0A0N0PBG2 A0A2H1VDI8 D6WM19 A0A2H1VF55 A0A0C9R581 A0A0J7N9Q1 A0A1B6FUI1 A0A158NLD7 A0A310SQL1 A0A2A3E268 K7JT31 A0A195B2A8 A0A0A9WIK4 A0A195CZP3 A0A0A9WB99 A0A0V0GCQ2 A0A232FB98 A0A0L7LHP3 A0A069DVT3 N6U6U8 A0A2A4JCH3 A0A0M9A4M1 A0A0L0CPS4 A0A0K8VAN7 A0A0K8V2C5 A0A0A1XDV5 A0A147AJZ0 A0A146Q6G1 A0A147AFX3 A0A146SBW7 A0A146RL18 A0A146Q4P7
A0A194PUN4 H9J196 A0A0N1IFZ7 A0A194PVA9 A0A2A4IYH7 A0A2H1VDP3 A0A0N0PBG2 A0A2H1VDI8 D6WM19 A0A2H1VF55 A0A0C9R581 A0A0J7N9Q1 A0A1B6FUI1 A0A158NLD7 A0A310SQL1 A0A2A3E268 K7JT31 A0A195B2A8 A0A0A9WIK4 A0A195CZP3 A0A0A9WB99 A0A0V0GCQ2 A0A232FB98 A0A0L7LHP3 A0A069DVT3 N6U6U8 A0A2A4JCH3 A0A0M9A4M1 A0A0L0CPS4 A0A0K8VAN7 A0A0K8V2C5 A0A0A1XDV5 A0A147AJZ0 A0A146Q6G1 A0A147AFX3 A0A146SBW7 A0A146RL18 A0A146Q4P7
PDB
5V3M
E-value=2.01075e-28,
Score=315
Ontologies
GO
Topology
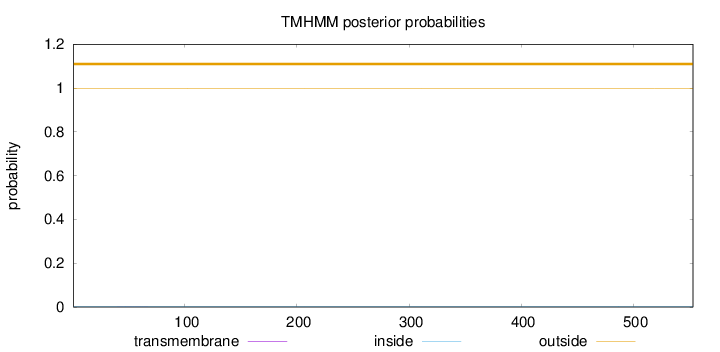
Length:
553
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01079
Exp number, first 60 AAs:
0.00643
Total prob of N-in:
0.00343
outside
1 - 553
Population Genetic Test Statistics
Pi
305.690778
Theta
223.156887
Tajima's D
1.172236
CLR
0
CSRT
0.707614619269037
Interpretation
Uncertain
