Gene
KWMTBOMO00682
Pre Gene Modal
BGIBMGA003217
Annotation
PREDICTED:_zinc_finger_protein_431-like_[Amyelois_transitella]
Full name
Zinc finger protein
Transcription factor
Location in the cell
Nuclear Reliability : 4.121
Sequence
CDS
ATGTACGAGAAGGAAAATATTCCCGAAATGACCGTTGAAGTCAAAACCGAATCGTTCGACATTGAAACCGAAGATCTATGCGAAGACGCCGATGTCGAGAGGAGGAACAACAATGAAGTTATCGATGATCTCAATGTTAATGAATTACAGATTACACCACAGCCGCTGGTTGACGTCTTTGTGTATTCGTGGAAATCGTACAAGTGGAAATGCTTACATTGTTCTGTTGCTTTTGACTCGATGAACGACTTGAGGAACCACAGTAAATTTGACCATAAGAAATGCTTTGGTTTTAAATGCGCAGACTGCGATGAGAGATTTAACACGTTCAGTACGTTTATCGAGCATGTTAGAGGGCATCGGGAAGACCTTAGAAATTACTGTCCGTATTGCAACAAGAGGTTCGACGACGCCGGCCAATGTATCGCGCATTTAACCGAGCACGGGATCAGCGAGGCCACAACCTGCCCCTACTGCGGCGAGATATTCCCTGCGACAGACCTCCTAGACACCCACAAGTCCATATACAAGATGGTTGAGATCCCTGAAGCCGTGACCGCAGCGCCGACCAAGGAGCGCCCTCCAAAAGTTGAGTACAAAATTCGCAAGGGCTGGATGAGGTATAAGTGGACTTGCCAGGATTGCTTCAAGGGCTTTGAAGGTGTCGTGCCTCTGAGGGCTCACGTGCAGAAAGAGCACAAGAAATGCTTCGCCATAAAGTGTATGGACTGCAACAGAAACATGAGGAACTATGCCAAGTTCGTAGAGCACGTCCACCAGCACCAGCCGTCTCTGAAAGAGTTCTGTCAGTTCTGCAACAAAAGAATTCCCAGCGAGAAGAAGTTGAAGGCCCACATCGAGGAGCACACGTCGGGTCCGCAGGCGCCGTGCTACGGCTGCGGACAGATCTTCGAAGACAACAAGATCTTGAAGAAGCACATGCTTGAATATGACGCCCCCGCGCCGCCCAAAGCGCTGCAAGGGCGGGACAGGTCCTGCGACCAGTGCGGCAAGGTGCTGGCCACGCCGGCCCGCCTCAGGACCCACAAGCTCACCCACGACCCTGACAGGCCGCGCAGCTTCATCTGCGACGTCTGCAACAAGGGCTTCCAGAGCAAAGAGAATTTGCGATGCCACAAGAGACTACACGGGACTAAGCGGCACGTTTGCCGGATCTGCCAGCGGGCCTTCTCCCTGCAGCTGGCGCTGCTGGAGCACCTGTCCAAGCACAGCTCGTTCAAGCAGTTCGAGTGCGACCACTGCGGCAAGAGGTTCTGGTCGAAGAACCGGCTGAGGCCGCACCTCAAGTGCCACGAGGAGGTCAAGCAGTTCGTCTGCAAGACCTGCGGGAAGGGGTTCAGGTTCAACAACCTGCTGGTCGCGCACGAGCGCCAGCACACGGGCACGAGGCCGCATGTGTGCGCGCTGTGCCGCCGCGCCTTCACCATCAGGCCGCTGCTGAACAAGCACATGGTGGCGGTGCACGGCACCACGCTGGTGCAGCACGAGGCAGCCACTAGGACCAGCAGGGACCAGGGGCGCTCCAAATAA
Protein
MYEKENIPEMTVEVKTESFDIETEDLCEDADVERRNNNEVIDDLNVNELQITPQPLVDVFVYSWKSYKWKCLHCSVAFDSMNDLRNHSKFDHKKCFGFKCADCDERFNTFSTFIEHVRGHREDLRNYCPYCNKRFDDAGQCIAHLTEHGISEATTCPYCGEIFPATDLLDTHKSIYKMVEIPEAVTAAPTKERPPKVEYKIRKGWMRYKWTCQDCFKGFEGVVPLRAHVQKEHKKCFAIKCMDCNRNMRNYAKFVEHVHQHQPSLKEFCQFCNKRIPSEKKLKAHIEEHTSGPQAPCYGCGQIFEDNKILKKHMLEYDAPAPPKALQGRDRSCDQCGKVLATPARLRTHKLTHDPDRPRSFICDVCNKGFQSKENLRCHKRLHGTKRHVCRICQRAFSLQLALLEHLSKHSSFKQFECDHCGKRFWSKNRLRPHLKCHEEVKQFVCKTCGKGFRFNNLLVAHERQHTGTRPHVCALCRRAFTIRPLLNKHMVAVHGTTLVQHEAATRTSRDQGRSK
Summary
Uniprot
H9J130
A0A2Y9R671
A0A3N0YPH5
A0A3P9PM12
E7F850
A0A0F8AD47
+ More
A0A2Y9QBG7 A0A3P9B4A9 A0A2J8JPS2 A0A147AHM6 A0A3P8T265 G1LM53 A0A3P8T3H0 A0A3P8SZU3 G1LM46 A0A286XWB0 H1A4D6 C3ZLR7 A0A3P8R268 A0A2Y9IAL6 F1Q8F4 A0A3P9B4C5 A0A3P8T4P5 A0A2Y9RB25 A0A3P9PEW8 G3UA26 A0A146PSZ5 A0A3B5KSK7 A0A2J8RFL1 A0A2U3ZAQ1 H9GQ93 A0A3B1JMP6 A0A3P9B4H1 A0A146N7G8 A0A287D9V8 A0A2K6LD93 A0A3P8R2L0 A0A3P8T0S7 A0A0L8IF58 F6XFC6 A0A146PGP9 A0A3B4GQQ8 S4RB94 A0A2K6PPK9 F1MUP0 Q3UP18 A0A0L8GY79 G3NKZ7 Q91WM0 A0A3B3W1H6 H9GTB2 A0A147AET0 I3KQE9 F1N5W2 A0A3B5R861 A0A3B5QXH2 A0A0D9QY95 A0A2K6PPL4 A0A2K6ADC7 Q9Z2X6 A3KP62 A0A146Z0C6 F6XDB9 A0A384DQW7 A0A2P6KF50 Q28E76 A0A2I3MWA2 A0A2K5IB44 A0A096M8D4 A0A2K6B6K7 F6ZJT8 A0A2I3M0J2 A0A2K6B6F9 A0A2K5VNE2 A0A2K6ADB6 A0A2K5IBE0 L8I0E0 A0A2K6B6G6 A0A0G2KA56 E7FDW1 A0A2K5VNB2 A0A2K6ADH3 F6UR85 A0A2K6B6E3 A0A384DRR4 A0A3N0YIY5
A0A2Y9QBG7 A0A3P9B4A9 A0A2J8JPS2 A0A147AHM6 A0A3P8T265 G1LM53 A0A3P8T3H0 A0A3P8SZU3 G1LM46 A0A286XWB0 H1A4D6 C3ZLR7 A0A3P8R268 A0A2Y9IAL6 F1Q8F4 A0A3P9B4C5 A0A3P8T4P5 A0A2Y9RB25 A0A3P9PEW8 G3UA26 A0A146PSZ5 A0A3B5KSK7 A0A2J8RFL1 A0A2U3ZAQ1 H9GQ93 A0A3B1JMP6 A0A3P9B4H1 A0A146N7G8 A0A287D9V8 A0A2K6LD93 A0A3P8R2L0 A0A3P8T0S7 A0A0L8IF58 F6XFC6 A0A146PGP9 A0A3B4GQQ8 S4RB94 A0A2K6PPK9 F1MUP0 Q3UP18 A0A0L8GY79 G3NKZ7 Q91WM0 A0A3B3W1H6 H9GTB2 A0A147AET0 I3KQE9 F1N5W2 A0A3B5R861 A0A3B5QXH2 A0A0D9QY95 A0A2K6PPL4 A0A2K6ADC7 Q9Z2X6 A3KP62 A0A146Z0C6 F6XDB9 A0A384DQW7 A0A2P6KF50 Q28E76 A0A2I3MWA2 A0A2K5IB44 A0A096M8D4 A0A2K6B6K7 F6ZJT8 A0A2I3M0J2 A0A2K6B6F9 A0A2K5VNE2 A0A2K6ADB6 A0A2K5IBE0 L8I0E0 A0A2K6B6G6 A0A0G2KA56 E7FDW1 A0A2K5VNB2 A0A2K6ADH3 F6UR85 A0A2K6B6E3 A0A384DRR4 A0A3N0YIY5
EC Number
2.1.1.-
Pubmed
EMBL
BABH01031481
RJVU01033420
ROL48109.1
FP102097
KQ043214
KKF09316.1
+ More
NBAG03000435 PNI24737.1 GCES01008253 JAR78070.1 ACTA01001889 ACTA01009889 ACTA01017889 ACTA01025889 ACTA01033889 ACTA01041888 ACTA01049888 ACTA01177880 ACTA01185878 ACTA01193876 AAKN02056734 ABQF01059460 ABQF01059461 ABQF01059462 ABQF01059463 ABQF01059464 ABQF01059465 ABQF01059466 ABQF01059467 ABQF01059468 ABQF01059469 GG666642 EEN46540.1 FP074867 GCES01139239 JAQ47083.1 NDHI03003702 PNJ07315.1 AAWZ02032867 GCES01158897 JAQ27425.1 AGTP01088332 KQ415854 KOG00078.1 GCES01143762 JAQ42560.1 AK143880 BAE25579.1 KQ419909 KOF82011.1 AC164531 BC014712 AAH14712.1 AAWZ02033925 GCES01009273 JAR77050.1 AERX01067313 AERX01067314 AERX01067315 AERX01067316 AQIB01141656 AB020542 BAA34724.1 BC134175 AAI34176.1 GCES01026868 JAR59455.1 JSUE03021256 MWRG01013681 PRD24960.1 CR848413 CAJ82437.1 AHZZ02013870 AHZZ02013871 AHZZ02013872 AHZZ02013873 AHZZ02013874 AHZZ02013875 AHZZ02013876 AHZZ02013877 AHZZ02013878 AYCK01011388 AQIA01034982 AQIA01034983 JH882314 ELR49950.1 AABR07009101 AABR07009102 AABR07009103 AABR07009104 AABR07009105 AABR07009106 RJVU01040460 ROL46202.1
NBAG03000435 PNI24737.1 GCES01008253 JAR78070.1 ACTA01001889 ACTA01009889 ACTA01017889 ACTA01025889 ACTA01033889 ACTA01041888 ACTA01049888 ACTA01177880 ACTA01185878 ACTA01193876 AAKN02056734 ABQF01059460 ABQF01059461 ABQF01059462 ABQF01059463 ABQF01059464 ABQF01059465 ABQF01059466 ABQF01059467 ABQF01059468 ABQF01059469 GG666642 EEN46540.1 FP074867 GCES01139239 JAQ47083.1 NDHI03003702 PNJ07315.1 AAWZ02032867 GCES01158897 JAQ27425.1 AGTP01088332 KQ415854 KOG00078.1 GCES01143762 JAQ42560.1 AK143880 BAE25579.1 KQ419909 KOF82011.1 AC164531 BC014712 AAH14712.1 AAWZ02033925 GCES01009273 JAR77050.1 AERX01067313 AERX01067314 AERX01067315 AERX01067316 AQIB01141656 AB020542 BAA34724.1 BC134175 AAI34176.1 GCES01026868 JAR59455.1 JSUE03021256 MWRG01013681 PRD24960.1 CR848413 CAJ82437.1 AHZZ02013870 AHZZ02013871 AHZZ02013872 AHZZ02013873 AHZZ02013874 AHZZ02013875 AHZZ02013876 AHZZ02013877 AHZZ02013878 AYCK01011388 AQIA01034982 AQIA01034983 JH882314 ELR49950.1 AABR07009101 AABR07009102 AABR07009103 AABR07009104 AABR07009105 AABR07009106 RJVU01040460 ROL46202.1
Proteomes
UP000005204
UP000248480
UP000242638
UP000000437
UP000265160
UP000265080
+ More
UP000008912 UP000005447 UP000007754 UP000001554 UP000265100 UP000248481 UP000007646 UP000261380 UP000245340 UP000001646 UP000018467 UP000005215 UP000233180 UP000053454 UP000002280 UP000261460 UP000245300 UP000233200 UP000009136 UP000007635 UP000000589 UP000261500 UP000005207 UP000002852 UP000029965 UP000233140 UP000006718 UP000261680 UP000028761 UP000233080 UP000028760 UP000233120 UP000233100 UP000002494
UP000008912 UP000005447 UP000007754 UP000001554 UP000265100 UP000248481 UP000007646 UP000261380 UP000245340 UP000001646 UP000018467 UP000005215 UP000233180 UP000053454 UP000002280 UP000261460 UP000245300 UP000233200 UP000009136 UP000007635 UP000000589 UP000261500 UP000005207 UP000002852 UP000029965 UP000233140 UP000006718 UP000261680 UP000028761 UP000233080 UP000028760 UP000233120 UP000233100 UP000002494
PRIDE
Interpro
ProteinModelPortal
H9J130
A0A2Y9R671
A0A3N0YPH5
A0A3P9PM12
E7F850
A0A0F8AD47
+ More
A0A2Y9QBG7 A0A3P9B4A9 A0A2J8JPS2 A0A147AHM6 A0A3P8T265 G1LM53 A0A3P8T3H0 A0A3P8SZU3 G1LM46 A0A286XWB0 H1A4D6 C3ZLR7 A0A3P8R268 A0A2Y9IAL6 F1Q8F4 A0A3P9B4C5 A0A3P8T4P5 A0A2Y9RB25 A0A3P9PEW8 G3UA26 A0A146PSZ5 A0A3B5KSK7 A0A2J8RFL1 A0A2U3ZAQ1 H9GQ93 A0A3B1JMP6 A0A3P9B4H1 A0A146N7G8 A0A287D9V8 A0A2K6LD93 A0A3P8R2L0 A0A3P8T0S7 A0A0L8IF58 F6XFC6 A0A146PGP9 A0A3B4GQQ8 S4RB94 A0A2K6PPK9 F1MUP0 Q3UP18 A0A0L8GY79 G3NKZ7 Q91WM0 A0A3B3W1H6 H9GTB2 A0A147AET0 I3KQE9 F1N5W2 A0A3B5R861 A0A3B5QXH2 A0A0D9QY95 A0A2K6PPL4 A0A2K6ADC7 Q9Z2X6 A3KP62 A0A146Z0C6 F6XDB9 A0A384DQW7 A0A2P6KF50 Q28E76 A0A2I3MWA2 A0A2K5IB44 A0A096M8D4 A0A2K6B6K7 F6ZJT8 A0A2I3M0J2 A0A2K6B6F9 A0A2K5VNE2 A0A2K6ADB6 A0A2K5IBE0 L8I0E0 A0A2K6B6G6 A0A0G2KA56 E7FDW1 A0A2K5VNB2 A0A2K6ADH3 F6UR85 A0A2K6B6E3 A0A384DRR4 A0A3N0YIY5
A0A2Y9QBG7 A0A3P9B4A9 A0A2J8JPS2 A0A147AHM6 A0A3P8T265 G1LM53 A0A3P8T3H0 A0A3P8SZU3 G1LM46 A0A286XWB0 H1A4D6 C3ZLR7 A0A3P8R268 A0A2Y9IAL6 F1Q8F4 A0A3P9B4C5 A0A3P8T4P5 A0A2Y9RB25 A0A3P9PEW8 G3UA26 A0A146PSZ5 A0A3B5KSK7 A0A2J8RFL1 A0A2U3ZAQ1 H9GQ93 A0A3B1JMP6 A0A3P9B4H1 A0A146N7G8 A0A287D9V8 A0A2K6LD93 A0A3P8R2L0 A0A3P8T0S7 A0A0L8IF58 F6XFC6 A0A146PGP9 A0A3B4GQQ8 S4RB94 A0A2K6PPK9 F1MUP0 Q3UP18 A0A0L8GY79 G3NKZ7 Q91WM0 A0A3B3W1H6 H9GTB2 A0A147AET0 I3KQE9 F1N5W2 A0A3B5R861 A0A3B5QXH2 A0A0D9QY95 A0A2K6PPL4 A0A2K6ADC7 Q9Z2X6 A3KP62 A0A146Z0C6 F6XDB9 A0A384DQW7 A0A2P6KF50 Q28E76 A0A2I3MWA2 A0A2K5IB44 A0A096M8D4 A0A2K6B6K7 F6ZJT8 A0A2I3M0J2 A0A2K6B6F9 A0A2K5VNE2 A0A2K6ADB6 A0A2K5IBE0 L8I0E0 A0A2K6B6G6 A0A0G2KA56 E7FDW1 A0A2K5VNB2 A0A2K6ADH3 F6UR85 A0A2K6B6E3 A0A384DRR4 A0A3N0YIY5
PDB
5V3M
E-value=6.392e-24,
Score=275
Ontologies
GO
Topology
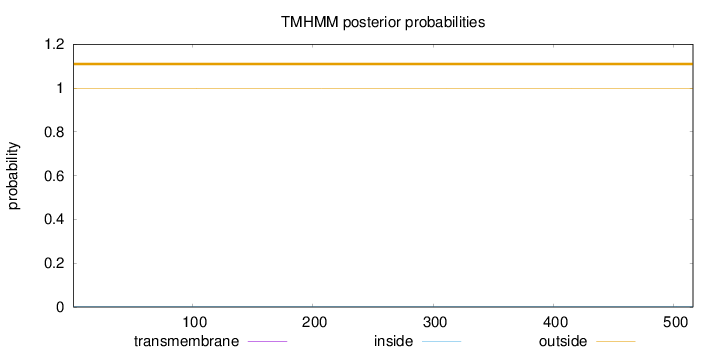
Length:
516
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00065
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00284
outside
1 - 516
Population Genetic Test Statistics
Pi
184.868281
Theta
170.240555
Tajima's D
1.251741
CLR
0.06206
CSRT
0.729313534323284
Interpretation
Uncertain
