Gene
KWMTBOMO00681
Pre Gene Modal
BGIBMGA003283
Annotation
PREDICTED:_gastrula_zinc_finger_protein_XlCGF26.1-like_[Amyelois_transitella]
Transcription factor
Location in the cell
Nuclear Reliability : 3.965
Sequence
CDS
ATGGAGAACATCGAAATATCATATTGCAGATTCTGCGCCGAATCTAAAAGCACCGATAAACTATTAAACTTACAAACAGACTGTCAGAAACACGAAGAAGTGCTCTTAAAACTGGCCTTTCTTAACGCTGTTTACGTTGACGTTTCTGACAAGAATTCACTACCGAAAACGGCTTGTTTAATATGTTACGAGTCTTTAAACAAAGCTTACGAGTTTTTACAAAGCGTCAAACAAGCCCAGGATGTTTTAATTGGCTTGTTTAGCACATCAGAGGACAACAAATGCGACCTATCGGACGACGATCAAAACTCCGCTTTCGATGATTTTCTTAGTTTAAACGAGTCTGTGGACATTAAATTAGAAACTATAGCCCAGGAGAATGAGTCGTTGGTTGATTCCGTCGAAGTCAAAGTGGAGCCAAAAGAAGAAGTTGAAACAATTAATCCAACGTTAGACGTTCATCAGATAATTGGGGCAGCCCTACAGGTCCCATTCGATCCGGAGATGACTATATACGCCAAGGATGTATCGGACGTAGGAACCAAGGCTATTAAGAGTTGGAGTCAGTACCCCTGGATCTGTTCACACTGTAATATAGAGTTTATCAACATAGACATGTTGAGGTCACACTCCAAATTAGTTCATGGCAGATGCTCAGCTCTGTTCTGTGTTGATTGTAAAATAGGAAGAAAGGACGACTTTATGGCTTTTGTGAAACACGTCAGAAAACATAGAAAGTGTTTGAGAAACTTCTGTCACTATTGTGACTCAGAGCTTCAAGGAGACGAAACAATTAGTAGTCATGTCAAGCAACACTTTCAGAAGTCTCAAATAGCGTGTCAACTGTGTGGTGAGATACTTAGTGATAAACACAATTTAGAAATACACCTACAGGACTACAGCTTGATGAAACCCAAACGTAAACCGAAGAGGAAACCAGGCACTGCCATCACTATTGAAGACTTAACCTGTAGTATATGTAAGAAAGTGTATAAGAACACAAACAGCCTACGAGATCACATGAAACTGCATAATATAAATCGTAAGAGAGACTACACTTGTGATCGGTGTGGGAAGATGTTCTATAATAAAGGTACGCTGACATCACACATTATGACACACGACAAGATACGTCCTCATATCTGTAGGATCTGCAACAGATCATTCCTCTTTCCAAATATGTTAAGAAGGCATGTGGAAATGCATTCTGGTGTGAAACCCTACTCTTGTGAACAATGCGGTCGATGCTTCAGACTACCTTATCAGTTGAACGCCCACAAAATAGTTCACACAGACTCAATGCCCTACATTTGTCAATATTGTAACAAAGCATTTAGATTTAAACAGATTTTGAAGAATCACGAGAGGCAACACACAGGCTCCAAGCCATATTCATGTCAGTTCTGTAGTATGGAATTCACAAACTGGTCAAATTACAATAAGCATATGAAGAGGAGACATGGCACTGATACAGCCAAAAAGAAAATCACTCCAGAGGGCGTTTTTCCTGTTGACCCCATCACTGGACAGATAGTACAAGTAGAAGACCCAGCCGGTACTGAGGAGTGGAAGAGTAAGATCATGATACCCGGGAAACGGGGGAAAAAGAAAAGAGTCAAACAAATTGACTGA
Protein
MENIEISYCRFCAESKSTDKLLNLQTDCQKHEEVLLKLAFLNAVYVDVSDKNSLPKTACLICYESLNKAYEFLQSVKQAQDVLIGLFSTSEDNKCDLSDDDQNSAFDDFLSLNESVDIKLETIAQENESLVDSVEVKVEPKEEVETINPTLDVHQIIGAALQVPFDPEMTIYAKDVSDVGTKAIKSWSQYPWICSHCNIEFINIDMLRSHSKLVHGRCSALFCVDCKIGRKDDFMAFVKHVRKHRKCLRNFCHYCDSELQGDETISSHVKQHFQKSQIACQLCGEILSDKHNLEIHLQDYSLMKPKRKPKRKPGTAITIEDLTCSICKKVYKNTNSLRDHMKLHNINRKRDYTCDRCGKMFYNKGTLTSHIMTHDKIRPHICRICNRSFLFPNMLRRHVEMHSGVKPYSCEQCGRCFRLPYQLNAHKIVHTDSMPYICQYCNKAFRFKQILKNHERQHTGSKPYSCQFCSMEFTNWSNYNKHMKRRHGTDTAKKKITPEGVFPVDPITGQIVQVEDPAGTEEWKSKIMIPGKRGKKKRVKQID
Summary
Uniprot
H9J196
A0A2H1VDP3
A0A0N1IFZ7
A0A194PUN4
A0A212FAD0
A0A1E1W4A4
+ More
A0A1E1W8Z0 A0A0L7LHY3 A0A2A4IYH7 A0A194PVA9 A0A2H1VE60 A0A2H1VDI8 A0A0N1IEG0 A0A0N1I7R1 A0A0N1IEE4 A0A2A4IW19 H9J139 D6WM19 A0A0L7L0W8 A0A1Y1KBP4 A0A1B6I4Q6 A0A1W4XD80 A0A069DVT3 N6U6U8 A0A0T6B7T6 A0A194PTE9 A0A0C9R581 A0A0A9XFI8 A0A2A3E268 A0A0L7LHP3 A0A0L7LDT9 A0A0A9WB99 A0A0A9WIK4 A0A310SQL1 A0A1B6FUI1 A0A023EZC1 A0A1B6CVD5 A0A067RQY6 A0A2J7QLG6 A0A3P8T3C0 A0A232F8A4 A0A146XBL3
A0A1E1W8Z0 A0A0L7LHY3 A0A2A4IYH7 A0A194PVA9 A0A2H1VE60 A0A2H1VDI8 A0A0N1IEG0 A0A0N1I7R1 A0A0N1IEE4 A0A2A4IW19 H9J139 D6WM19 A0A0L7L0W8 A0A1Y1KBP4 A0A1B6I4Q6 A0A1W4XD80 A0A069DVT3 N6U6U8 A0A0T6B7T6 A0A194PTE9 A0A0C9R581 A0A0A9XFI8 A0A2A3E268 A0A0L7LHP3 A0A0L7LDT9 A0A0A9WB99 A0A0A9WIK4 A0A310SQL1 A0A1B6FUI1 A0A023EZC1 A0A1B6CVD5 A0A067RQY6 A0A2J7QLG6 A0A3P8T3C0 A0A232F8A4 A0A146XBL3
Pubmed
EMBL
BABH01031482
ODYU01001969
SOQ38896.1
KQ460940
KPJ10603.1
KQ459593
+ More
KPI96693.1 AGBW02009496 OWR50687.1 GDQN01009343 JAT81711.1 GDQN01007620 JAT83434.1 JTDY01001068 KOB74974.1 NWSH01005474 PCG64173.1 KPI96694.1 ODYU01001823 SOQ38554.1 SOQ38895.1 KPJ10633.1 KPJ10606.1 KPJ10605.1 NWSH01005726 PCG64005.1 BABH01031441 KQ971343 EFA04207.1 JTDY01003842 KOB68924.1 GEZM01091329 JAV56906.1 GECU01025790 JAS81916.1 GBGD01001082 JAC87807.1 APGK01037606 KB740948 ENN77375.1 LJIG01009280 KRT83396.1 KPI96696.1 GBYB01002000 GBYB01002001 JAG71767.1 JAG71768.1 GBHO01024870 GBHO01024869 GDHC01016743 JAG18734.1 JAG18735.1 JAQ01886.1 KZ288430 PBC25833.1 KOB74972.1 JTDY01001547 KOB73570.1 GBHO01038932 GBHO01038928 GBHO01038927 GBHO01025725 GDHC01010384 JAG04672.1 JAG04676.1 JAG04677.1 JAG17879.1 JAQ08245.1 GBHO01038929 GBHO01025727 GBHO01019226 GBHO01011035 GDHC01017998 JAG04675.1 JAG17877.1 JAG24378.1 JAG32569.1 JAQ00631.1 KQ760118 OAD61926.1 GECZ01015909 JAS53860.1 GBBI01004137 JAC14575.1 GEDC01020003 GEDC01013481 JAS17295.1 JAS23817.1 KK852521 KDR22139.1 NEVH01013247 PNF29417.1 NNAY01000782 OXU26537.1 GCES01046795 JAR39528.1
KPI96693.1 AGBW02009496 OWR50687.1 GDQN01009343 JAT81711.1 GDQN01007620 JAT83434.1 JTDY01001068 KOB74974.1 NWSH01005474 PCG64173.1 KPI96694.1 ODYU01001823 SOQ38554.1 SOQ38895.1 KPJ10633.1 KPJ10606.1 KPJ10605.1 NWSH01005726 PCG64005.1 BABH01031441 KQ971343 EFA04207.1 JTDY01003842 KOB68924.1 GEZM01091329 JAV56906.1 GECU01025790 JAS81916.1 GBGD01001082 JAC87807.1 APGK01037606 KB740948 ENN77375.1 LJIG01009280 KRT83396.1 KPI96696.1 GBYB01002000 GBYB01002001 JAG71767.1 JAG71768.1 GBHO01024870 GBHO01024869 GDHC01016743 JAG18734.1 JAG18735.1 JAQ01886.1 KZ288430 PBC25833.1 KOB74972.1 JTDY01001547 KOB73570.1 GBHO01038932 GBHO01038928 GBHO01038927 GBHO01025725 GDHC01010384 JAG04672.1 JAG04676.1 JAG04677.1 JAG17879.1 JAQ08245.1 GBHO01038929 GBHO01025727 GBHO01019226 GBHO01011035 GDHC01017998 JAG04675.1 JAG17877.1 JAG24378.1 JAG32569.1 JAQ00631.1 KQ760118 OAD61926.1 GECZ01015909 JAS53860.1 GBBI01004137 JAC14575.1 GEDC01020003 GEDC01013481 JAS17295.1 JAS23817.1 KK852521 KDR22139.1 NEVH01013247 PNF29417.1 NNAY01000782 OXU26537.1 GCES01046795 JAR39528.1
Proteomes
Interpro
ProteinModelPortal
H9J196
A0A2H1VDP3
A0A0N1IFZ7
A0A194PUN4
A0A212FAD0
A0A1E1W4A4
+ More
A0A1E1W8Z0 A0A0L7LHY3 A0A2A4IYH7 A0A194PVA9 A0A2H1VE60 A0A2H1VDI8 A0A0N1IEG0 A0A0N1I7R1 A0A0N1IEE4 A0A2A4IW19 H9J139 D6WM19 A0A0L7L0W8 A0A1Y1KBP4 A0A1B6I4Q6 A0A1W4XD80 A0A069DVT3 N6U6U8 A0A0T6B7T6 A0A194PTE9 A0A0C9R581 A0A0A9XFI8 A0A2A3E268 A0A0L7LHP3 A0A0L7LDT9 A0A0A9WB99 A0A0A9WIK4 A0A310SQL1 A0A1B6FUI1 A0A023EZC1 A0A1B6CVD5 A0A067RQY6 A0A2J7QLG6 A0A3P8T3C0 A0A232F8A4 A0A146XBL3
A0A1E1W8Z0 A0A0L7LHY3 A0A2A4IYH7 A0A194PVA9 A0A2H1VE60 A0A2H1VDI8 A0A0N1IEG0 A0A0N1I7R1 A0A0N1IEE4 A0A2A4IW19 H9J139 D6WM19 A0A0L7L0W8 A0A1Y1KBP4 A0A1B6I4Q6 A0A1W4XD80 A0A069DVT3 N6U6U8 A0A0T6B7T6 A0A194PTE9 A0A0C9R581 A0A0A9XFI8 A0A2A3E268 A0A0L7LHP3 A0A0L7LDT9 A0A0A9WB99 A0A0A9WIK4 A0A310SQL1 A0A1B6FUI1 A0A023EZC1 A0A1B6CVD5 A0A067RQY6 A0A2J7QLG6 A0A3P8T3C0 A0A232F8A4 A0A146XBL3
PDB
5V3G
E-value=3.35284e-27,
Score=304
Ontologies
GO
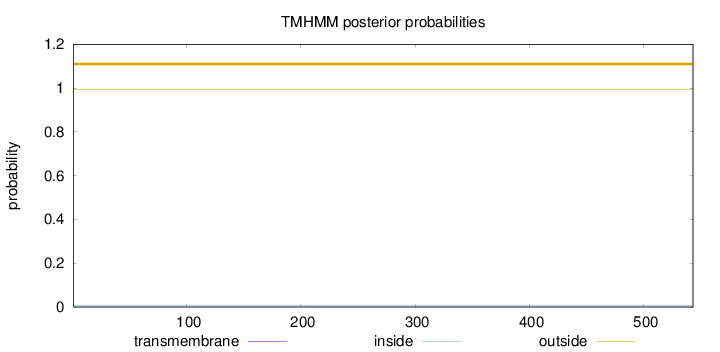
Topology
Length:
543
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00049
Exp number, first 60 AAs:
0.00049
Total prob of N-in:
0.00524
outside
1 - 543
Population Genetic Test Statistics
Pi
251.889307
Theta
223.95819
Tajima's D
0.322806
CLR
0.15978
CSRT
0.462476876156192
Interpretation
Uncertain
