Gene
KWMTBOMO00654
Pre Gene Modal
BGIBMGA003870
Annotation
PREDICTED:_cycle_like_factor_b_isoform_X1_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 3.893
Sequence
CDS
ATGCCGAAGGAGATCGATGAGCGGGGCTACCTCAGGATGACGCAGGACTGTGCGAACCCGAGTGCGATACAGGCGTATGAGATGACACCGGAAGGGGGCGTGGGGTTGGGCGGCGCGTGCGCCGACTCGGCCGGCGCCCTCATCACCCCGCACCCTCCCCTTCATCATCCAGTGCCGCAGACCTCACAGCAACTCCATCATGATCCGCGGAAAACAAAACCCAATCACTACGTTCCTGAAAACTATGAGATATCGGCGTGCGACTCGCAGCGCCAGTCTCCACACGGACACACGCCGCGGACTCGGACCAACAGTACTCGTAAACGGAAACCATCGTCCTATGGAACTGGAAGCGCTTATGATGACGACGAAGAAGATTCAAGATCCACAACAACTACAACCACGGCGACCACGCGCGGCACTCCCGACAAGAAACAAAATCATAGCGAGATCGAAAAACGGCGACGTGATAAAATGAACACTTTTATATCGGAACTGAGCGCCATGATACCGATGTGCGGAGCCATGGCTCGGAAGCTGGACAAGCTGACAGTCCTCCGCATGGCCGTTCAACACTTGAGGACGGTGCGCGGCGCGCTCTCGGCCTGCCCCTTGACGGCCCGGCCCTGTCCCACTTACCTCACTGAGCGAGAACTAAACGCACTGATACTCCAGGCGGCTCACGACTGCTTTTTGCTCGTCGTGGGATGCGATCGCGGCAGGTTACTTTACGTCTCGGCTTCGGTCAAAAATATACTTCATTACGATCAATCGGAACTATTAGGCCAGAGCCTGTTCGACATTTTACATCCAAAGGACGTCGCTAAAGTAAAGGAACAACTATCATCGTCAGACCTTAGTCCACGAGAAAGACTCATCGACGCAAAAACGATGTTGCCATTGAAAGCGGACGTAGTAGCCGGCGCGTCCAGGTTCGGTCCGGGGGCACGTCGGTCGTTCTTCTGTCGGATCAAGTGCAAATTAGACACAGAAGAAGTCGAAACTCCCCCGCAGCCAGTCAAGGAGGAGGTGGAGCCAGTGGCCAAGATGCGGAAGAAACACTCTCACGAGAAAAAATACTGCGTCGTGCAATGCACAGGTTATTTAAAATCGTGGGCGCCAACGAAGATGTGTGACGGTGCCAGTGCGGAAGGGGGAGAGGAGAGTGAAGCGTGCAACATGTCGTGTCTAGTGGCAGTGGGCCGAACCTTGGGTGGTTTAGCTCCCACCACGAACTCTCCGACATCCATGCCTCAAACCAGACACCTGCAATACGTCTCACGACACACTACCGACGGGAAATTCTTGTTTGTTGATCAGCGGGTGACTCTCGCGCTAGGATTCTTGCCCCAGGAGTTGCTGGGCACTAGTCTGTACGAGTACGTGCACGGGCCGGAGCTGGGCGCCGTGGCCCGCACGCACAAGGCCGCGCTGCTGCAGCGCGACGCGCTCCACACCCCGCCCTACTGCTTCCGCAGGAAGAACGGCTCGATGGCCAGAATACAAACCCACTTCAAACCGTTCAAAAATCCGTGGACGAAAGACGTCGAGTGCTTAGTGGCCAACAACACCGTCGTGTCGGAGTCCCAGGTGTCGCTGCAGCAAGACACGACGCAAGCAGCGTTTGATATTTACAAGCAAAAATCGGACGTCGAGATGCAGCGCCTCATAGACTCGCAAGTCGAGTCCCATAAGATCGGCAGCGCCATCGCCGAGGAGGCTCTGCGGCGGTCGTCGACTGATTACTCGCCCGACCTGCCGACCGAGTTGCTGCAAGACGCCGTCTTCAATCAGCAGTTTTCTCCGCTGCAAGTTGCGCTAGTGGACAACATCCTCGGGACCGACAGTAGCACTTCAAATCAGGTTCGCAACAACGTGCCCCTGAGTTCAGTGAGCTCCATCAGCCCGCCGGCGCCGTCGGTGGAAGAGACGACTCTGTGCGACACCCCCCCGCCGCCTTCCCCCCCGCTGCCTTCCCCCCCGCTGCCGCCCCTGGTCATGGACGGCAATGGTGAAGCAGCAATGGCAGTTATCATGAGTCTTCTAGAAGCGGACGCAGGTCTCGGAGGGCCGATCAATTTTTCAGGTCTTCCTTGGCCTTTGCCCTGA
Protein
MPKEIDERGYLRMTQDCANPSAIQAYEMTPEGGVGLGGACADSAGALITPHPPLHHPVPQTSQQLHHDPRKTKPNHYVPENYEISACDSQRQSPHGHTPRTRTNSTRKRKPSSYGTGSAYDDDEEDSRSTTTTTTATTRGTPDKKQNHSEIEKRRRDKMNTFISELSAMIPMCGAMARKLDKLTVLRMAVQHLRTVRGALSACPLTARPCPTYLTERELNALILQAAHDCFLLVVGCDRGRLLYVSASVKNILHYDQSELLGQSLFDILHPKDVAKVKEQLSSSDLSPRERLIDAKTMLPLKADVVAGASRFGPGARRSFFCRIKCKLDTEEVETPPQPVKEEVEPVAKMRKKHSHEKKYCVVQCTGYLKSWAPTKMCDGASAEGGEESEACNMSCLVAVGRTLGGLAPTTNSPTSMPQTRHLQYVSRHTTDGKFLFVDQRVTLALGFLPQELLGTSLYEYVHGPELGAVARTHKAALLQRDALHTPPYCFRRKNGSMARIQTHFKPFKNPWTKDVECLVANNTVVSESQVSLQQDTTQAAFDIYKQKSDVEMQRLIDSQVESHKIGSAIAEEALRRSSTDYSPDLPTELLQDAVFNQQFSPLQVALVDNILGTDSSTSNQVRNNVPLSSVSSISPPAPSVEETTLCDTPPPPSPPLPSPPLPPLVMDGNGEAAMAVIMSLLEADAGLGGPINFSGLPWPLP
Summary
Uniprot
Q8N0R5
H9J2Y3
A0A2S1ZDB6
A0A2A4JQ56
A0A1U9XQN3
A0A2W1BM79
+ More
A0A2S0DDS3 Q3ZTR4 A0A212EKE6 A0A194QYE8 Q6VRU5 Q2PRP1 A0A0F7IGS0 A9XCF1 A0A2J7Q3V6 A0A1L8DM25 A0A1L8DMD6 Q05J77 A0A0M8ZQ63 K7IW03 E2A3F0 A0MH07 E2B7K2 A0A087ZYH3 A0A026W1Y8 A0A151IRV0 A0A1J1IIJ1 A0A195EX82 A0A1Y1NEK2 A0A151WT58 A0A195BTR2 A0A3L8DNB8 A0A158NNS1 A0A1B0DPF6 A1IM63 A0A154P0Z1 A0A1J1ILV8 T1H956 E0W1Z5 A0A310SFD3 A0A1L5YQJ1 K4F2M9 A0A084W480 Q17JB2 Q17JB1 A0A1Q3FQ15 A0A1Q3FPV7 A0A1Q3FPW8 A0A1Q3FPQ4 A0A1Q3FPM4 A0A1B0YZI3 U5EXM2 A0A146LWP6 A0A286Q1C4 A0A2I6BPZ8 A0A146LYC2 T2C8X8 A0A2S0X9U9 U5NDP5 A0A2P6KD60 M4HZ37 A0A2T7PK89
A0A2S0DDS3 Q3ZTR4 A0A212EKE6 A0A194QYE8 Q6VRU5 Q2PRP1 A0A0F7IGS0 A9XCF1 A0A2J7Q3V6 A0A1L8DM25 A0A1L8DMD6 Q05J77 A0A0M8ZQ63 K7IW03 E2A3F0 A0MH07 E2B7K2 A0A087ZYH3 A0A026W1Y8 A0A151IRV0 A0A1J1IIJ1 A0A195EX82 A0A1Y1NEK2 A0A151WT58 A0A195BTR2 A0A3L8DNB8 A0A158NNS1 A0A1B0DPF6 A1IM63 A0A154P0Z1 A0A1J1ILV8 T1H956 E0W1Z5 A0A310SFD3 A0A1L5YQJ1 K4F2M9 A0A084W480 Q17JB2 Q17JB1 A0A1Q3FQ15 A0A1Q3FPV7 A0A1Q3FPW8 A0A1Q3FPQ4 A0A1Q3FPM4 A0A1B0YZI3 U5EXM2 A0A146LWP6 A0A286Q1C4 A0A2I6BPZ8 A0A146LYC2 T2C8X8 A0A2S0X9U9 U5NDP5 A0A2P6KD60 M4HZ37 A0A2T7PK89
Pubmed
EMBL
AB073712
BAB91178.1
BABH01031193
BABH01031194
BABH01031195
BABH01031196
+ More
BABH01031197 MF374804 AWK22768.1 NWSH01000799 PCG74131.1 KY682108 KY825136 AQZ26709.1 AVE16219.1 KZ150031 PZC74724.1 KY940303 ASK39990.1 AY364478 AAR13012.1 AGBW02014274 OWR41954.1 KQ460953 KPJ10324.1 AY330487 AAR14937.1 DQ305400 ABC18168.2 KP147946 AKG92783.1 EF174304 ABO86538.1 NEVH01018428 PNF23249.1 GFDF01006561 JAV07523.1 GFDF01006560 JAV07524.1 AB278116 BAF35030.1 KQ435912 KOX68860.1 GL436428 EFN72014.1 DQ841151 ABI21880.1 GL446181 EFN88377.1 KK107485 EZA50085.1 KQ981124 KYN09334.1 CVRI01000054 CRL00055.1 KQ981953 KYN32489.1 GEZM01009202 JAV94626.1 KQ982766 KYQ50981.1 KQ976417 KYM89821.1 QOIP01000006 RLU21940.1 ADTU01021739 ADTU01021740 ADTU01021741 ADTU01021742 AJVK01008090 AJVK01008091 AB289690 BAF44540.1 KQ434785 KZC05014.1 CRL00054.1 ACPB03010320 ACPB03010321 ACPB03010322 DS235873 EEB19589.1 KQ766175 OAD53726.1 KX602315 APP94024.1 JN573265 AEX32872.1 ATLV01020285 KE525297 KFB45024.1 CH477233 EAT46793.1 EAT46794.1 GFDL01005421 JAV29624.1 GFDL01005527 JAV29518.1 GFDL01005493 JAV29552.1 GFDL01005553 JAV29492.1 GFDL01005528 JAV29517.1 KX014725 ANO53974.1 GANO01000885 JAB58986.1 GDHC01007030 JAQ11599.1 KX238952 ANW48377.1 KY922996 AUI80367.1 GDHC01006035 JAQ12594.1 KC885968 AGV28715.1 MG280774 MF175153 AWC08576.1 AXF35705.1 KF316926 AGX93014.1 MWRG01015605 PRD24247.1 JQ670886 AFV39705.1 PZQS01000003 PVD33849.1
BABH01031197 MF374804 AWK22768.1 NWSH01000799 PCG74131.1 KY682108 KY825136 AQZ26709.1 AVE16219.1 KZ150031 PZC74724.1 KY940303 ASK39990.1 AY364478 AAR13012.1 AGBW02014274 OWR41954.1 KQ460953 KPJ10324.1 AY330487 AAR14937.1 DQ305400 ABC18168.2 KP147946 AKG92783.1 EF174304 ABO86538.1 NEVH01018428 PNF23249.1 GFDF01006561 JAV07523.1 GFDF01006560 JAV07524.1 AB278116 BAF35030.1 KQ435912 KOX68860.1 GL436428 EFN72014.1 DQ841151 ABI21880.1 GL446181 EFN88377.1 KK107485 EZA50085.1 KQ981124 KYN09334.1 CVRI01000054 CRL00055.1 KQ981953 KYN32489.1 GEZM01009202 JAV94626.1 KQ982766 KYQ50981.1 KQ976417 KYM89821.1 QOIP01000006 RLU21940.1 ADTU01021739 ADTU01021740 ADTU01021741 ADTU01021742 AJVK01008090 AJVK01008091 AB289690 BAF44540.1 KQ434785 KZC05014.1 CRL00054.1 ACPB03010320 ACPB03010321 ACPB03010322 DS235873 EEB19589.1 KQ766175 OAD53726.1 KX602315 APP94024.1 JN573265 AEX32872.1 ATLV01020285 KE525297 KFB45024.1 CH477233 EAT46793.1 EAT46794.1 GFDL01005421 JAV29624.1 GFDL01005527 JAV29518.1 GFDL01005493 JAV29552.1 GFDL01005553 JAV29492.1 GFDL01005528 JAV29517.1 KX014725 ANO53974.1 GANO01000885 JAB58986.1 GDHC01007030 JAQ11599.1 KX238952 ANW48377.1 KY922996 AUI80367.1 GDHC01006035 JAQ12594.1 KC885968 AGV28715.1 MG280774 MF175153 AWC08576.1 AXF35705.1 KF316926 AGX93014.1 MWRG01015605 PRD24247.1 JQ670886 AFV39705.1 PZQS01000003 PVD33849.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000235965
UP000053105
+ More
UP000002358 UP000000311 UP000008237 UP000005203 UP000053097 UP000078492 UP000183832 UP000078541 UP000075809 UP000078540 UP000279307 UP000005205 UP000092462 UP000076502 UP000015103 UP000009046 UP000030765 UP000008820 UP000245119
UP000002358 UP000000311 UP000008237 UP000005203 UP000053097 UP000078492 UP000183832 UP000078541 UP000075809 UP000078540 UP000279307 UP000005205 UP000092462 UP000076502 UP000015103 UP000009046 UP000030765 UP000008820 UP000245119
Interpro
Gene 3D
ProteinModelPortal
Q8N0R5
H9J2Y3
A0A2S1ZDB6
A0A2A4JQ56
A0A1U9XQN3
A0A2W1BM79
+ More
A0A2S0DDS3 Q3ZTR4 A0A212EKE6 A0A194QYE8 Q6VRU5 Q2PRP1 A0A0F7IGS0 A9XCF1 A0A2J7Q3V6 A0A1L8DM25 A0A1L8DMD6 Q05J77 A0A0M8ZQ63 K7IW03 E2A3F0 A0MH07 E2B7K2 A0A087ZYH3 A0A026W1Y8 A0A151IRV0 A0A1J1IIJ1 A0A195EX82 A0A1Y1NEK2 A0A151WT58 A0A195BTR2 A0A3L8DNB8 A0A158NNS1 A0A1B0DPF6 A1IM63 A0A154P0Z1 A0A1J1ILV8 T1H956 E0W1Z5 A0A310SFD3 A0A1L5YQJ1 K4F2M9 A0A084W480 Q17JB2 Q17JB1 A0A1Q3FQ15 A0A1Q3FPV7 A0A1Q3FPW8 A0A1Q3FPQ4 A0A1Q3FPM4 A0A1B0YZI3 U5EXM2 A0A146LWP6 A0A286Q1C4 A0A2I6BPZ8 A0A146LYC2 T2C8X8 A0A2S0X9U9 U5NDP5 A0A2P6KD60 M4HZ37 A0A2T7PK89
A0A2S0DDS3 Q3ZTR4 A0A212EKE6 A0A194QYE8 Q6VRU5 Q2PRP1 A0A0F7IGS0 A9XCF1 A0A2J7Q3V6 A0A1L8DM25 A0A1L8DMD6 Q05J77 A0A0M8ZQ63 K7IW03 E2A3F0 A0MH07 E2B7K2 A0A087ZYH3 A0A026W1Y8 A0A151IRV0 A0A1J1IIJ1 A0A195EX82 A0A1Y1NEK2 A0A151WT58 A0A195BTR2 A0A3L8DNB8 A0A158NNS1 A0A1B0DPF6 A1IM63 A0A154P0Z1 A0A1J1ILV8 T1H956 E0W1Z5 A0A310SFD3 A0A1L5YQJ1 K4F2M9 A0A084W480 Q17JB2 Q17JB1 A0A1Q3FQ15 A0A1Q3FPV7 A0A1Q3FPW8 A0A1Q3FPQ4 A0A1Q3FPM4 A0A1B0YZI3 U5EXM2 A0A146LWP6 A0A286Q1C4 A0A2I6BPZ8 A0A146LYC2 T2C8X8 A0A2S0X9U9 U5NDP5 A0A2P6KD60 M4HZ37 A0A2T7PK89
PDB
4F3L
E-value=1.62029e-105,
Score=980
Ontologies
GO
Topology
Subcellular location
Nucleus
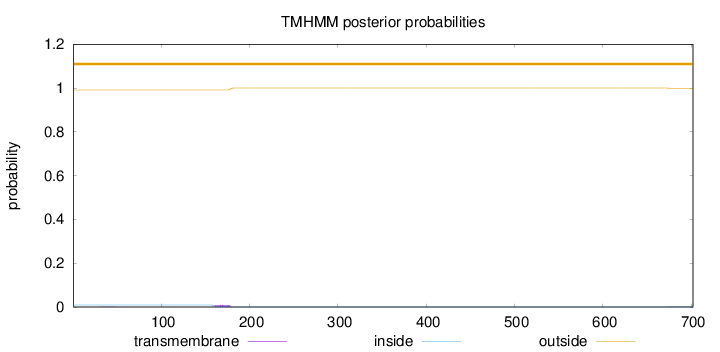
Length:
702
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.17508
Exp number, first 60 AAs:
0.00345
Total prob of N-in:
0.00902
outside
1 - 702
Population Genetic Test Statistics
Pi
20.20687
Theta
16.359408
Tajima's D
-1.3018
CLR
0.19702
CSRT
0.0841957902104895
Interpretation
Uncertain
