Gene
KWMTBOMO00089
Pre Gene Modal
BGIBMGA002019
Annotation
PREDICTED:_deformed_epidermal_autoregulatory_factor_1_isoform_X1_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 4.388
Sequence
CDS
ATGGCCGAGAATAGAAGTTCGGACAACGTCGTCATGCCGGACATTACCGAAGTATCCGAAGCCGATCCATTGTCGACGGCGACCGAGCACGACAACGAGCGGGTGGAGGGCACGTCTGTGTTGTCTACCACCATCAAAAACACCCGGCTGTCCAATGATTCTAACGGAGTTGTGACTGTACCAGTCTCGTTGCCAGTTGGGACGCTCATCGCGGGTACCACCTTCAACGTAATCACGTCAGACCATTTGCCGCATTTCAAGCCAATGATCTGCGTTGACAATGGTTTTATTTCGGGTGGGCCAGTGAGCGATGAGTTAAAAGCAACACACATCGTCATCCAGAACCCTCCTTCTCCTACTCGAACTAATTGCGAGGATGTGGTGGTAGCTGCGCCTGCGCCGCGATCGATGAGTGCACGATCATGGACGGAGACTGCGAACATGCCAGTGCTGCCGGTACGCTGTAAAAATACATCAGCAGAGCTCCACAAAAATAAATTCGGTTCAGGGGGCAGAGGCAGGTGTATCAAATACGGCATAGAGTGGTACACTCCTAGTGAGTTCGAAGCTCTATGTGGCAGAGCCTCTAGCAAAGATTGGAAGAGGTCTATAAGGTTTGGTGGAAGAAGTATTCAAGCTCTCATTGATGAGGGTATTTTGACTCCACATGCTACAAGCTGCACTTGCGGAGCTTGCTGTGATGATAAATCTGCCATGGGACCAGTGAGACTATTCACTCCATATAAAAGAAAGCGAAAGGAAGAAAGAAGTGAGAAGAAACCAAAAATTAAAAGAGAAAGCAGCATTGGAGAGAGTGAAGTGGATAACATACACCAGAGCAGTGAGAATAGTCACTCAAAAGAAGCTTGGCAGAGCATCGCTGAGGGTTTGGAAACTACTACCGATTACCAGATACTGGGGAACCCTGAAATCAAGACTGACCCCCCTCCAACAGATGTACCAGACATGAGCGCTGTTTTAAAACGTCTAGATGACATCAGTGAAACCCTAGTGAGGCTGTCCACGGAACTAAAGCAATGTGTAGAAGATGTTAGACGGGTCAGTGCTCGACAACTAGAGCGCTTGGAGCAAGAACGGACGTCAGCCCTACTCGCTGCCAGTGTCGATGATCAGGTTGATCCTGAGCAAGTATTATTGCATAATGTTGAAGATACAGAATCTAAGAAGTGCGCTAATTGCAACCGTGAAGCATCAGCAGAATGTTCTTTATGCAGGCGTACACCATATTGTTCCACATATTGTCAGAAGAAGGACTGGGCTTCACATCAGATTGAGTGCCTACGATCGGTGCCAGCAATACATACTGATGCACAACAGCACCAATCAATAATGCTGATTGTTGAAAGCCAGCACCAATAG
Protein
MAENRSSDNVVMPDITEVSEADPLSTATEHDNERVEGTSVLSTTIKNTRLSNDSNGVVTVPVSLPVGTLIAGTTFNVITSDHLPHFKPMICVDNGFISGGPVSDELKATHIVIQNPPSPTRTNCEDVVVAAPAPRSMSARSWTETANMPVLPVRCKNTSAELHKNKFGSGGRGRCIKYGIEWYTPSEFEALCGRASSKDWKRSIRFGGRSIQALIDEGILTPHATSCTCGACCDDKSAMGPVRLFTPYKRKRKEERSEKKPKIKRESSIGESEVDNIHQSSENSHSKEAWQSIAEGLETTTDYQILGNPEIKTDPPPTDVPDMSAVLKRLDDISETLVRLSTELKQCVEDVRRVSARQLERLEQERTSALLAASVDDQVDPEQVLLHNVEDTESKKCANCNREASAECSLCRRTPYCSTYCQKKDWASHQIECLRSVPAIHTDAQQHQSIMLIVESQHQ
Summary
Uniprot
H9IXN6
A0A212F9L0
A0A2A4J853
A0A194Q4T4
A0A194QZR4
A0A2W1BND9
+ More
D6WIQ4 A0A1W4WX63 A0A1Y1MCZ0 N6U7X6 J3JTD8 A0A0T6BI46 A0A195FTZ6 A0A158NNI0 A0A151X7S9 A0A151I3R2 E2A6U5 A0A195C1N3 A0A2A3EPJ8 A0A087ZS71 A0A0L7RD56 A0A1B0GJX6 A0A154PK68 A0A026W2K1 A0A310SG00 A0A0N0U6Y5 A0A1L8DNC5 K7J9U9 E2C3D0 A0A0L0C639 A0A0A9YCZ0 A0A1B6CZD8 A0A1B6HPP0 A0A084WKX2 A0A0K8V169 A0A0K8UPY3 A0A034WQT7 W8C687 A0A1I8PNQ0 A0A0A1WYM9 A0A1I8MC44 T1PBZ6 A0A3L8D5A3 A0A1B0FC71 A0A1A9UXW6 A0A1A9YN18 A0A1B0BAB9 A0A1A9Z3K4 A0A1A9WS91 A0A0J7KA85 U5EQX5 A0A2H8TG45 E0W221 J9L485 A0A151ITJ7 A0A2S2QWN6 A0A2R7WM81 J9JZJ0 A0A1B6LDB1 J9KVF6
D6WIQ4 A0A1W4WX63 A0A1Y1MCZ0 N6U7X6 J3JTD8 A0A0T6BI46 A0A195FTZ6 A0A158NNI0 A0A151X7S9 A0A151I3R2 E2A6U5 A0A195C1N3 A0A2A3EPJ8 A0A087ZS71 A0A0L7RD56 A0A1B0GJX6 A0A154PK68 A0A026W2K1 A0A310SG00 A0A0N0U6Y5 A0A1L8DNC5 K7J9U9 E2C3D0 A0A0L0C639 A0A0A9YCZ0 A0A1B6CZD8 A0A1B6HPP0 A0A084WKX2 A0A0K8V169 A0A0K8UPY3 A0A034WQT7 W8C687 A0A1I8PNQ0 A0A0A1WYM9 A0A1I8MC44 T1PBZ6 A0A3L8D5A3 A0A1B0FC71 A0A1A9UXW6 A0A1A9YN18 A0A1B0BAB9 A0A1A9Z3K4 A0A1A9WS91 A0A0J7KA85 U5EQX5 A0A2H8TG45 E0W221 J9L485 A0A151ITJ7 A0A2S2QWN6 A0A2R7WM81 J9JZJ0 A0A1B6LDB1 J9KVF6
Pubmed
EMBL
BABH01015789
BABH01015790
AGBW02009564
OWR50424.1
NWSH01002716
PCG67630.1
+ More
KQ459463 KPJ00548.1 KQ461108 KPJ09081.1 KZ149952 PZC76569.1 KQ971321 EEZ99506.1 GEZM01038200 JAV81726.1 APGK01039325 KB740969 KB632230 ENN76751.1 ERL90391.1 BT126494 AEE61458.1 LJIG01000373 KRT86567.1 KQ981268 KYN43901.1 ADTU01002719 ADTU01002720 ADTU01002721 KQ982431 KYQ56435.1 KQ976492 KYM83300.1 GL437203 EFN70811.1 KQ978344 KYM94784.1 KZ288204 PBC33192.1 KQ414615 KOC68749.1 AJWK01022434 KQ434931 KZC11854.1 KK107462 QOIP01000013 EZA50315.1 RLU15194.1 KQ760382 OAD60707.1 KQ435714 KOX79202.1 GFDF01006220 JAV07864.1 AAZX01008588 GL452320 EFN77468.1 JRES01000836 KNC27863.1 GBHO01014631 GDHC01018559 JAG28973.1 JAQ00070.1 GEDC01018645 JAS18653.1 GECU01031141 JAS76565.1 ATLV01024156 KE525350 KFB50866.1 GDHF01019776 JAI32538.1 GDHF01023580 JAI28734.1 GAKP01002397 JAC56555.1 GAMC01001262 JAC05294.1 GBXI01010546 JAD03746.1 KA646291 AFP60920.1 RLU15193.1 CCAG010016073 JXJN01010895 LBMM01010817 KMQ87212.1 GANO01003075 JAB56796.1 GFXV01001281 MBW13086.1 DS235873 EEB19615.1 ABLF02029098 ABLF02029103 KQ981001 KYN10414.1 GGMS01012974 MBY82177.1 KK855086 PTY20737.1 ABLF02020605 ABLF02020607 GEBQ01018316 JAT21661.1 ABLF02016786
KQ459463 KPJ00548.1 KQ461108 KPJ09081.1 KZ149952 PZC76569.1 KQ971321 EEZ99506.1 GEZM01038200 JAV81726.1 APGK01039325 KB740969 KB632230 ENN76751.1 ERL90391.1 BT126494 AEE61458.1 LJIG01000373 KRT86567.1 KQ981268 KYN43901.1 ADTU01002719 ADTU01002720 ADTU01002721 KQ982431 KYQ56435.1 KQ976492 KYM83300.1 GL437203 EFN70811.1 KQ978344 KYM94784.1 KZ288204 PBC33192.1 KQ414615 KOC68749.1 AJWK01022434 KQ434931 KZC11854.1 KK107462 QOIP01000013 EZA50315.1 RLU15194.1 KQ760382 OAD60707.1 KQ435714 KOX79202.1 GFDF01006220 JAV07864.1 AAZX01008588 GL452320 EFN77468.1 JRES01000836 KNC27863.1 GBHO01014631 GDHC01018559 JAG28973.1 JAQ00070.1 GEDC01018645 JAS18653.1 GECU01031141 JAS76565.1 ATLV01024156 KE525350 KFB50866.1 GDHF01019776 JAI32538.1 GDHF01023580 JAI28734.1 GAKP01002397 JAC56555.1 GAMC01001262 JAC05294.1 GBXI01010546 JAD03746.1 KA646291 AFP60920.1 RLU15193.1 CCAG010016073 JXJN01010895 LBMM01010817 KMQ87212.1 GANO01003075 JAB56796.1 GFXV01001281 MBW13086.1 DS235873 EEB19615.1 ABLF02029098 ABLF02029103 KQ981001 KYN10414.1 GGMS01012974 MBY82177.1 KK855086 PTY20737.1 ABLF02020605 ABLF02020607 GEBQ01018316 JAT21661.1 ABLF02016786
Proteomes
UP000005204
UP000007151
UP000218220
UP000053268
UP000053240
UP000007266
+ More
UP000192223 UP000019118 UP000030742 UP000078541 UP000005205 UP000075809 UP000078540 UP000000311 UP000078542 UP000242457 UP000005203 UP000053825 UP000092461 UP000076502 UP000053097 UP000279307 UP000053105 UP000002358 UP000008237 UP000037069 UP000030765 UP000095300 UP000095301 UP000092444 UP000078200 UP000092443 UP000092460 UP000092445 UP000091820 UP000036403 UP000009046 UP000007819 UP000078492
UP000192223 UP000019118 UP000030742 UP000078541 UP000005205 UP000075809 UP000078540 UP000000311 UP000078542 UP000242457 UP000005203 UP000053825 UP000092461 UP000076502 UP000053097 UP000279307 UP000053105 UP000002358 UP000008237 UP000037069 UP000030765 UP000095300 UP000095301 UP000092444 UP000078200 UP000092443 UP000092460 UP000092445 UP000091820 UP000036403 UP000009046 UP000007819 UP000078492
PRIDE
Interpro
SUPFAM
SSF63763
SSF63763
Gene 3D
ProteinModelPortal
H9IXN6
A0A212F9L0
A0A2A4J853
A0A194Q4T4
A0A194QZR4
A0A2W1BND9
+ More
D6WIQ4 A0A1W4WX63 A0A1Y1MCZ0 N6U7X6 J3JTD8 A0A0T6BI46 A0A195FTZ6 A0A158NNI0 A0A151X7S9 A0A151I3R2 E2A6U5 A0A195C1N3 A0A2A3EPJ8 A0A087ZS71 A0A0L7RD56 A0A1B0GJX6 A0A154PK68 A0A026W2K1 A0A310SG00 A0A0N0U6Y5 A0A1L8DNC5 K7J9U9 E2C3D0 A0A0L0C639 A0A0A9YCZ0 A0A1B6CZD8 A0A1B6HPP0 A0A084WKX2 A0A0K8V169 A0A0K8UPY3 A0A034WQT7 W8C687 A0A1I8PNQ0 A0A0A1WYM9 A0A1I8MC44 T1PBZ6 A0A3L8D5A3 A0A1B0FC71 A0A1A9UXW6 A0A1A9YN18 A0A1B0BAB9 A0A1A9Z3K4 A0A1A9WS91 A0A0J7KA85 U5EQX5 A0A2H8TG45 E0W221 J9L485 A0A151ITJ7 A0A2S2QWN6 A0A2R7WM81 J9JZJ0 A0A1B6LDB1 J9KVF6
D6WIQ4 A0A1W4WX63 A0A1Y1MCZ0 N6U7X6 J3JTD8 A0A0T6BI46 A0A195FTZ6 A0A158NNI0 A0A151X7S9 A0A151I3R2 E2A6U5 A0A195C1N3 A0A2A3EPJ8 A0A087ZS71 A0A0L7RD56 A0A1B0GJX6 A0A154PK68 A0A026W2K1 A0A310SG00 A0A0N0U6Y5 A0A1L8DNC5 K7J9U9 E2C3D0 A0A0L0C639 A0A0A9YCZ0 A0A1B6CZD8 A0A1B6HPP0 A0A084WKX2 A0A0K8V169 A0A0K8UPY3 A0A034WQT7 W8C687 A0A1I8PNQ0 A0A0A1WYM9 A0A1I8MC44 T1PBZ6 A0A3L8D5A3 A0A1B0FC71 A0A1A9UXW6 A0A1A9YN18 A0A1B0BAB9 A0A1A9Z3K4 A0A1A9WS91 A0A0J7KA85 U5EQX5 A0A2H8TG45 E0W221 J9L485 A0A151ITJ7 A0A2S2QWN6 A0A2R7WM81 J9JZJ0 A0A1B6LDB1 J9KVF6
PDB
1H5P
E-value=6.63595e-08,
Score=137
Ontologies
GO
PANTHER
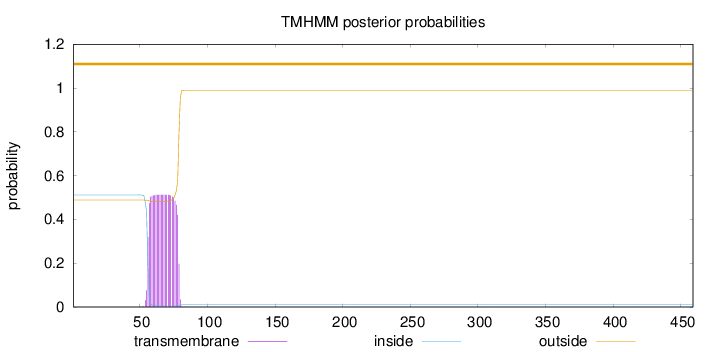
Topology
Length:
459
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
11.68215
Exp number, first 60 AAs:
2.42156
Total prob of N-in:
0.51188
outside
1 - 459
Population Genetic Test Statistics
Pi
21.257879
Theta
23.337689
Tajima's D
0.151904
CLR
0.007177
CSRT
0.415679216039198
Interpretation
Uncertain
