Gene
KWMTBOMO16873
Pre Gene Modal
BGIBMGA012644
Annotation
PREDICTED:_ras-related_protein_Rab-23-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.266 Cytoplasmic Reliability : 2.266
Sequence
CDS
ATGCGCGAGGAGGAGCTGGAGGTGGCCCTGAAGGTGGTGATCGTCGGTGACGGCGGAGTCGGCAAGTCCAGTATGATACAGCGCTACTGCCGCGGCACCTTCACCAGGGACTACAAGAAGACCATCGGGGTCGACTTTCTTGAGAGACAGATCGAGATAGACGGCGAGGAAGTCCGACTGATGCTGTGGGACACGGCGGGCCAGGAGGAGTTCGACGCGATCACCAAGGCGTACTACCGCGGCGCCCACGCCTGCGTGCTCGCCTTCTCCACAACCGACCGAGACTCTTTCTTAGCGCTGCACTCCTGGAAGTTGAAAGTAAGTCTGTCTTATGCTCGGATGAATACAGTAGTAGTTCTTGACAACAGTGATCCGTGA
Protein
MREEELEVALKVVIVGDGGVGKSSMIQRYCRGTFTRDYKKTIGVDFLERQIEIDGEEVRLMLWDTAGQEEFDAITKAYYRGAHACVLAFSTTDRDSFLALHSWKLKVSLSYARMNTVVVLDNSDP
Summary
Uniprot
H9JSY2
A0A194QF45
A0A212F6T1
A0A194QX90
A0A437BM64
A0A2W1BVI8
+ More
A0A222AIZ2 D6WRN4 A0A0L7LIH6 J3JWQ1 A0A1Y1M3N4 C4WRN6 A0A2S2QFN0 A0A2J7QER9 A0A067R0K1 A0A2H1VHP0 A0A146M0M0 A0A1B6IGI0 A0A1B6KRG3 A0A1B6GRJ4 A0A1W4X3Q8 A0A2S2PD02 A0A0P4VMA0 R4G8N7 A0A023F8Z6 A0A165VFY4 A0A0A9YTJ0 K7J5H1 A0A3Q0IQI9 E2AZR4 A0A195CBJ6 F4WY88 A0A195AX51 E2B7Q7 A0A158NHS1 A0A3L8D8F4 A0A2A3E5B8 K1QR50 U5EVL0 V5FYB7 V4B4A1 A0A1S3IEG9 A0A3L8SZK5 A0A232EWF1 A0A034WM63 A0A3B4A1H8 A0A1B6JTP9 A0A0K8U3Q5 A0A3B5LX33 A0A2M4BYK0 Q17KZ8 A0A182GYX6 A0A0A1WKS7 A0A2M4CLT6 A0A2M4CLZ3 A0A2M4AWL3 A0A2M4CLX6 A0A2M4CWX9 H0ZRC9 A0A3N0XTE7 A0A182P027 A0A182WES8 A0A182UX90 A0A182TUU9 A0A182LP37 A0A182X2N4 A0A182HN94 Q7QGJ4 A0A182LRA3 A0A182RY17 A0A182MXS1 A0A182QPU5 A0A1Q3FKN0 A0A182Y606 B0X036 W5JLP2 A0A1I8MPK8 A0A182FU70 A0A3Q0SUN0 A0A1A9ZZZ2 A0A182JC48 A0A091EWC5 U3JG74 B8A453 Q503R5 A0A1A9YCT5 A0A0L0CM89 I3JTP9 A0A3M0L115 A0A1B0BUG5 A0A1J1I7H6 A0A1A9V7X4 A0A1B0G9L8 A0A1I8P1C7 A8VIF4 A0A146Y9R8 T1IX75 A0A3Q3M9G5 A0A3B3XIN4 A0A093BVE0
A0A222AIZ2 D6WRN4 A0A0L7LIH6 J3JWQ1 A0A1Y1M3N4 C4WRN6 A0A2S2QFN0 A0A2J7QER9 A0A067R0K1 A0A2H1VHP0 A0A146M0M0 A0A1B6IGI0 A0A1B6KRG3 A0A1B6GRJ4 A0A1W4X3Q8 A0A2S2PD02 A0A0P4VMA0 R4G8N7 A0A023F8Z6 A0A165VFY4 A0A0A9YTJ0 K7J5H1 A0A3Q0IQI9 E2AZR4 A0A195CBJ6 F4WY88 A0A195AX51 E2B7Q7 A0A158NHS1 A0A3L8D8F4 A0A2A3E5B8 K1QR50 U5EVL0 V5FYB7 V4B4A1 A0A1S3IEG9 A0A3L8SZK5 A0A232EWF1 A0A034WM63 A0A3B4A1H8 A0A1B6JTP9 A0A0K8U3Q5 A0A3B5LX33 A0A2M4BYK0 Q17KZ8 A0A182GYX6 A0A0A1WKS7 A0A2M4CLT6 A0A2M4CLZ3 A0A2M4AWL3 A0A2M4CLX6 A0A2M4CWX9 H0ZRC9 A0A3N0XTE7 A0A182P027 A0A182WES8 A0A182UX90 A0A182TUU9 A0A182LP37 A0A182X2N4 A0A182HN94 Q7QGJ4 A0A182LRA3 A0A182RY17 A0A182MXS1 A0A182QPU5 A0A1Q3FKN0 A0A182Y606 B0X036 W5JLP2 A0A1I8MPK8 A0A182FU70 A0A3Q0SUN0 A0A1A9ZZZ2 A0A182JC48 A0A091EWC5 U3JG74 B8A453 Q503R5 A0A1A9YCT5 A0A0L0CM89 I3JTP9 A0A3M0L115 A0A1B0BUG5 A0A1J1I7H6 A0A1A9V7X4 A0A1B0G9L8 A0A1I8P1C7 A8VIF4 A0A146Y9R8 T1IX75 A0A3Q3M9G5 A0A3B3XIN4 A0A093BVE0
Pubmed
19121390
26354079
22118469
28756777
18362917
19820115
+ More
26227816 22516182 23537049 28004739 24845553 26823975 27129103 25474469 25401762 20075255 20798317 21719571 21347285 30249741 22992520 23254933 30282656 28648823 25348373 25463417 17510324 26483478 25830018 20360741 20966253 12364791 14747013 17210077 25244985 20920257 23761445 25315136 23594743 26108605 25186727 17937392
26227816 22516182 23537049 28004739 24845553 26823975 27129103 25474469 25401762 20075255 20798317 21719571 21347285 30249741 22992520 23254933 30282656 28648823 25348373 25463417 17510324 26483478 25830018 20360741 20966253 12364791 14747013 17210077 25244985 20920257 23761445 25315136 23594743 26108605 25186727 17937392
EMBL
BABH01038906
BABH01038907
KQ459053
KPJ04039.1
AGBW02009975
OWR49440.1
+ More
KQ461073 KPJ09570.1 RSAL01000034 RVE51415.1 KZ149982 PZC75773.1 KY563815 ASO76338.1 KQ971354 EFA07061.1 JTDY01000937 KOB75368.1 APGK01036247 APGK01036248 APGK01036249 APGK01036250 BT127669 KB740937 KB631668 AEE62631.1 ENN77764.1 ERL85155.1 GEZM01041973 JAV80081.1 ABLF02031457 AK339845 BAH70556.1 GGMS01007345 MBY76548.1 NEVH01015306 PNF27081.1 KK853035 KDR12233.1 ODYU01002616 SOQ40345.1 GDHC01005650 JAQ12979.1 GECU01021676 JAS86030.1 GEBQ01025935 JAT14042.1 GECZ01004740 JAS65029.1 GGMR01014708 MBY27327.1 GDKW01000346 JAI56249.1 ACPB03020901 GAHY01000652 JAA76858.1 GBBI01000787 JAC17925.1 KT984801 AMZ00353.1 GBHO01008668 GBRD01010339 GDHC01018430 JAG34936.1 JAG55485.1 JAQ00199.1 GL444277 EFN61043.1 KQ978023 KYM98070.1 GL888439 EGI60845.1 KQ976725 KYM76622.1 GL446201 EFN88270.1 ADTU01016033 ADTU01016034 QOIP01000011 RLU16765.1 KZ288386 PBC26482.1 JH817566 EKC36223.1 GANO01000976 JAB58895.1 GALX01005796 JAB62670.1 KB199650 ESP05283.1 QUSF01000002 RLW12354.1 NNAY01001882 OXU22651.1 GAKP01003500 JAC55452.1 GECU01005167 JAT02540.1 GDHF01032539 GDHF01031349 GDHF01021756 JAI19775.1 JAI20965.1 JAI30558.1 GGFJ01009024 MBW58165.1 CH477220 EAT47357.1 JXUM01021594 KQ560606 KXJ81643.1 GBXI01014648 JAC99643.1 GGFL01002124 MBW66302.1 GGFL01002125 MBW66303.1 GGFK01011862 MBW45183.1 GGFL01002158 MBW66336.1 GGFL01005658 MBW69836.1 ABQF01001020 RJVU01061435 ROJ36049.1 APCN01001891 AAAB01008834 EAA05694.4 AXCM01000524 AXCN02002308 GFDL01007002 JAV28043.1 DS232228 EDS37911.1 ADMH02000695 ETN65287.1 KK719242 KFO62183.1 AGTO01010618 BX004843 BC095213 BC164825 AAH95213.1 AAI64825.1 JRES01000301 KNC32579.1 AERX01006384 AERX01006385 QRBI01000099 RMC17474.1 JXJN01020691 CVRI01000042 CRK95532.1 CCAG010022856 EU176872 ABW24032.1 GCES01035444 JAR50879.1 JH431644 KL229652 KFV04834.1
KQ461073 KPJ09570.1 RSAL01000034 RVE51415.1 KZ149982 PZC75773.1 KY563815 ASO76338.1 KQ971354 EFA07061.1 JTDY01000937 KOB75368.1 APGK01036247 APGK01036248 APGK01036249 APGK01036250 BT127669 KB740937 KB631668 AEE62631.1 ENN77764.1 ERL85155.1 GEZM01041973 JAV80081.1 ABLF02031457 AK339845 BAH70556.1 GGMS01007345 MBY76548.1 NEVH01015306 PNF27081.1 KK853035 KDR12233.1 ODYU01002616 SOQ40345.1 GDHC01005650 JAQ12979.1 GECU01021676 JAS86030.1 GEBQ01025935 JAT14042.1 GECZ01004740 JAS65029.1 GGMR01014708 MBY27327.1 GDKW01000346 JAI56249.1 ACPB03020901 GAHY01000652 JAA76858.1 GBBI01000787 JAC17925.1 KT984801 AMZ00353.1 GBHO01008668 GBRD01010339 GDHC01018430 JAG34936.1 JAG55485.1 JAQ00199.1 GL444277 EFN61043.1 KQ978023 KYM98070.1 GL888439 EGI60845.1 KQ976725 KYM76622.1 GL446201 EFN88270.1 ADTU01016033 ADTU01016034 QOIP01000011 RLU16765.1 KZ288386 PBC26482.1 JH817566 EKC36223.1 GANO01000976 JAB58895.1 GALX01005796 JAB62670.1 KB199650 ESP05283.1 QUSF01000002 RLW12354.1 NNAY01001882 OXU22651.1 GAKP01003500 JAC55452.1 GECU01005167 JAT02540.1 GDHF01032539 GDHF01031349 GDHF01021756 JAI19775.1 JAI20965.1 JAI30558.1 GGFJ01009024 MBW58165.1 CH477220 EAT47357.1 JXUM01021594 KQ560606 KXJ81643.1 GBXI01014648 JAC99643.1 GGFL01002124 MBW66302.1 GGFL01002125 MBW66303.1 GGFK01011862 MBW45183.1 GGFL01002158 MBW66336.1 GGFL01005658 MBW69836.1 ABQF01001020 RJVU01061435 ROJ36049.1 APCN01001891 AAAB01008834 EAA05694.4 AXCM01000524 AXCN02002308 GFDL01007002 JAV28043.1 DS232228 EDS37911.1 ADMH02000695 ETN65287.1 KK719242 KFO62183.1 AGTO01010618 BX004843 BC095213 BC164825 AAH95213.1 AAI64825.1 JRES01000301 KNC32579.1 AERX01006384 AERX01006385 QRBI01000099 RMC17474.1 JXJN01020691 CVRI01000042 CRK95532.1 CCAG010022856 EU176872 ABW24032.1 GCES01035444 JAR50879.1 JH431644 KL229652 KFV04834.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000053240
UP000283053
UP000007266
+ More
UP000037510 UP000019118 UP000030742 UP000007819 UP000235965 UP000027135 UP000192223 UP000015103 UP000002358 UP000079169 UP000000311 UP000078542 UP000007755 UP000078540 UP000008237 UP000005205 UP000279307 UP000242457 UP000005408 UP000030746 UP000085678 UP000276834 UP000215335 UP000261520 UP000261380 UP000008820 UP000069940 UP000249989 UP000007754 UP000075885 UP000075920 UP000075903 UP000075902 UP000075882 UP000076407 UP000075840 UP000007062 UP000075883 UP000075900 UP000075884 UP000075886 UP000076408 UP000002320 UP000000673 UP000095301 UP000069272 UP000261340 UP000092445 UP000075880 UP000052976 UP000016665 UP000000437 UP000092443 UP000037069 UP000005207 UP000269221 UP000092460 UP000183832 UP000078200 UP000092444 UP000095300 UP000265000 UP000261660 UP000261480
UP000037510 UP000019118 UP000030742 UP000007819 UP000235965 UP000027135 UP000192223 UP000015103 UP000002358 UP000079169 UP000000311 UP000078542 UP000007755 UP000078540 UP000008237 UP000005205 UP000279307 UP000242457 UP000005408 UP000030746 UP000085678 UP000276834 UP000215335 UP000261520 UP000261380 UP000008820 UP000069940 UP000249989 UP000007754 UP000075885 UP000075920 UP000075903 UP000075902 UP000075882 UP000076407 UP000075840 UP000007062 UP000075883 UP000075900 UP000075884 UP000075886 UP000076408 UP000002320 UP000000673 UP000095301 UP000069272 UP000261340 UP000092445 UP000075880 UP000052976 UP000016665 UP000000437 UP000092443 UP000037069 UP000005207 UP000269221 UP000092460 UP000183832 UP000078200 UP000092444 UP000095300 UP000265000 UP000261660 UP000261480
PRIDE
Pfam
PF00071 Ras
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
H9JSY2
A0A194QF45
A0A212F6T1
A0A194QX90
A0A437BM64
A0A2W1BVI8
+ More
A0A222AIZ2 D6WRN4 A0A0L7LIH6 J3JWQ1 A0A1Y1M3N4 C4WRN6 A0A2S2QFN0 A0A2J7QER9 A0A067R0K1 A0A2H1VHP0 A0A146M0M0 A0A1B6IGI0 A0A1B6KRG3 A0A1B6GRJ4 A0A1W4X3Q8 A0A2S2PD02 A0A0P4VMA0 R4G8N7 A0A023F8Z6 A0A165VFY4 A0A0A9YTJ0 K7J5H1 A0A3Q0IQI9 E2AZR4 A0A195CBJ6 F4WY88 A0A195AX51 E2B7Q7 A0A158NHS1 A0A3L8D8F4 A0A2A3E5B8 K1QR50 U5EVL0 V5FYB7 V4B4A1 A0A1S3IEG9 A0A3L8SZK5 A0A232EWF1 A0A034WM63 A0A3B4A1H8 A0A1B6JTP9 A0A0K8U3Q5 A0A3B5LX33 A0A2M4BYK0 Q17KZ8 A0A182GYX6 A0A0A1WKS7 A0A2M4CLT6 A0A2M4CLZ3 A0A2M4AWL3 A0A2M4CLX6 A0A2M4CWX9 H0ZRC9 A0A3N0XTE7 A0A182P027 A0A182WES8 A0A182UX90 A0A182TUU9 A0A182LP37 A0A182X2N4 A0A182HN94 Q7QGJ4 A0A182LRA3 A0A182RY17 A0A182MXS1 A0A182QPU5 A0A1Q3FKN0 A0A182Y606 B0X036 W5JLP2 A0A1I8MPK8 A0A182FU70 A0A3Q0SUN0 A0A1A9ZZZ2 A0A182JC48 A0A091EWC5 U3JG74 B8A453 Q503R5 A0A1A9YCT5 A0A0L0CM89 I3JTP9 A0A3M0L115 A0A1B0BUG5 A0A1J1I7H6 A0A1A9V7X4 A0A1B0G9L8 A0A1I8P1C7 A8VIF4 A0A146Y9R8 T1IX75 A0A3Q3M9G5 A0A3B3XIN4 A0A093BVE0
A0A222AIZ2 D6WRN4 A0A0L7LIH6 J3JWQ1 A0A1Y1M3N4 C4WRN6 A0A2S2QFN0 A0A2J7QER9 A0A067R0K1 A0A2H1VHP0 A0A146M0M0 A0A1B6IGI0 A0A1B6KRG3 A0A1B6GRJ4 A0A1W4X3Q8 A0A2S2PD02 A0A0P4VMA0 R4G8N7 A0A023F8Z6 A0A165VFY4 A0A0A9YTJ0 K7J5H1 A0A3Q0IQI9 E2AZR4 A0A195CBJ6 F4WY88 A0A195AX51 E2B7Q7 A0A158NHS1 A0A3L8D8F4 A0A2A3E5B8 K1QR50 U5EVL0 V5FYB7 V4B4A1 A0A1S3IEG9 A0A3L8SZK5 A0A232EWF1 A0A034WM63 A0A3B4A1H8 A0A1B6JTP9 A0A0K8U3Q5 A0A3B5LX33 A0A2M4BYK0 Q17KZ8 A0A182GYX6 A0A0A1WKS7 A0A2M4CLT6 A0A2M4CLZ3 A0A2M4AWL3 A0A2M4CLX6 A0A2M4CWX9 H0ZRC9 A0A3N0XTE7 A0A182P027 A0A182WES8 A0A182UX90 A0A182TUU9 A0A182LP37 A0A182X2N4 A0A182HN94 Q7QGJ4 A0A182LRA3 A0A182RY17 A0A182MXS1 A0A182QPU5 A0A1Q3FKN0 A0A182Y606 B0X036 W5JLP2 A0A1I8MPK8 A0A182FU70 A0A3Q0SUN0 A0A1A9ZZZ2 A0A182JC48 A0A091EWC5 U3JG74 B8A453 Q503R5 A0A1A9YCT5 A0A0L0CM89 I3JTP9 A0A3M0L115 A0A1B0BUG5 A0A1J1I7H6 A0A1A9V7X4 A0A1B0G9L8 A0A1I8P1C7 A8VIF4 A0A146Y9R8 T1IX75 A0A3Q3M9G5 A0A3B3XIN4 A0A093BVE0
PDB
1Z2A
E-value=1.95795e-37,
Score=384
Ontologies
GO
PANTHER
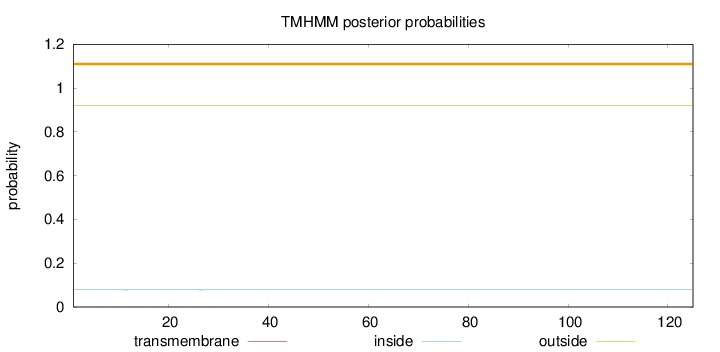
Topology
Length:
125
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01215
Exp number, first 60 AAs:
0.01013
Total prob of N-in:
0.07867
outside
1 - 125
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
0
CSRT
0
Interpretation
Uncertain
